7OUL
 
 | | BDM88832 inhibitor bound to the transmembrane domain of AcrB-R971A | | Descriptor: | (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, 1,2-ETHANEDIOL, 1-(3-chloranyl-5-iodanyl-pyridin-2-yl)piperazine, ... | | Authors: | Tam, H.K, Foong, W.E, Pos, K.M. | | Deposit date: | 2021-06-12 | | Release date: | 2021-12-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Pyridylpiperazine-based allosteric inhibitors of RND-type multidrug efflux pumps.
Nat Commun, 13, 2022
|
|
7OUK
 
 | | BDM88855 inhibitor bound to the transmembrane domain of AcrB | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl didecanoate, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, 1,2-ETHANEDIOL, ... | | Authors: | Tam, H.K, Foong, W.E, Pos, K.M. | | Deposit date: | 2021-06-12 | | Release date: | 2021-12-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Pyridylpiperazine-based allosteric inhibitors of RND-type multidrug efflux pumps.
Nat Commun, 13, 2022
|
|
7OUM
 
 | | BDM88855 inhibitor bound to the transmembrane domain of AcrB-R971A | | Descriptor: | (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, 1,2-ETHANEDIOL, 3-chloranyl-2-piperazin-1-yl-quinoline, ... | | Authors: | Tam, H.K, Foong, W.E, Pos, K.M. | | Deposit date: | 2021-06-12 | | Release date: | 2021-12-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Pyridylpiperazine-based allosteric inhibitors of RND-type multidrug efflux pumps.
Nat Commun, 13, 2022
|
|
5FNR
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | (3S)-3-(4-chlorophenyl)-3-(1-methylbenzotriazol-5-yl)propanoic acid, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
5FNQ
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | 3-(4-CHLOROPHENYL)PROPANOIC ACID, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
5FNU
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | (3S)-3-(7-methoxy-1-methyl-1H-benzo[d][1,2,3]triazol-5-yl)-3-(4-methyl-3-(((R)-4-methyl-1,1-dioxido-3,4-dihydro-2H-benzo[b][1,4,5]oxathiazepin-2-yl)methyl)phenyl)propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
5FZJ
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | 2,6-DIMETHYL-4H-PYRANO[3,4-D][1,3]OXAZOL-4-ONE, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2016-03-14 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
5FNS
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | (3s)-{4-Chloro-3-[(N-methylmethanesulfonamido) methyl]phenyl}-3-(1-methyl-1H-1,2,3-benzotriazol-5-yl) propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
5FNT
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | (3S)-3-{4-Chloro-3-[(N-methylbenzenesulfonamido) methyl]phenyl}-3-(1-methyl-1H-1,2,3-benzotriazol-5-yl)propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
5FZN
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | KELCH-LIKE ECH-ASSOCIATED PROTEIN 1, SULFATE ION, benzenesulfonamide | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2016-03-15 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
7QB2
 
 | | Pim1 in complex with (E)-4-((6-amino-1-methyl-2-oxoindolin-3-ylidene)methyl)benzoic acid and Pimtide | | Descriptor: | 4-[(E)-(6-azanyl-1-methyl-2-oxidanylidene-indol-3-ylidene)methyl]benzoic acid, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2021-11-18 | | Release date: | 2022-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
7QFM
 
 | | Pim1 in complex with (E)-4-((2-oxoindolin-3-ylidene)methyl)benzoic acid and Pimtide | | Descriptor: | 4-[(~{E})-(2-oxidanylidene-1~{H}-indol-3-ylidene)methyl]benzoic acid, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
2KYA
 
 | | Solution structure of the leader sequence of the patellamide precursor peptide, PatE1-34 | | Descriptor: | Patellamide protein | | Authors: | Houssen, W.E, Wright, S.H, Kalverda, A.P, Thompson, G.S, Kelly, S.M, Jaspars, M. | | Deposit date: | 2010-05-21 | | Release date: | 2010-09-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Leader Sequence of the Patellamide Precursor Peptide, PatE(1-34).
Chembiochem, 11, 2010
|
|
2MCX
 
 | | Solid-state NMR structure of piscidin 3 in aligned 1:1 phosphatidylethanolamine/phosphoglycerol lipid bilayers | | Descriptor: | Piscidin-3 | | Authors: | Fu, R, Tian, Y, Perrin Jr, B.S, Grant, C.V, Wieczorek, W.E, Pastor, R.W, Cotten, M.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-01-22 | | Last modified: | 2021-08-18 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution structures and orientations of antimicrobial peptides piscidin 1 and piscidin 3 in fluid bilayers reveal tilting, kinking, and bilayer immersion.
J.Am.Chem.Soc., 136, 2014
|
|
2OIP
 
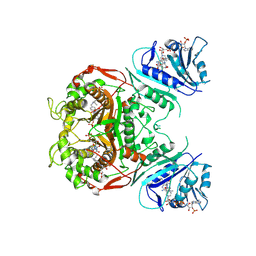 | |
4HRR
 
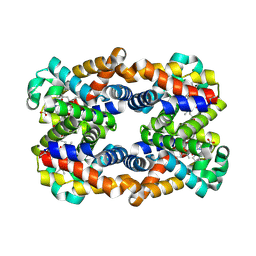 | | Scapharca tetrameric hemoglobin, CO-state | | Descriptor: | CARBON MONOXIDE, Globin-2 A chain, Hemoglobin B chain, ... | | Authors: | Royer, W.E. | | Deposit date: | 2012-10-28 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Tertiary and Quaternary Allostery in Tetrameric Hemoglobin from Scapharca inaequivalvis.
Biochemistry, 52, 2013
|
|
1JWN
 
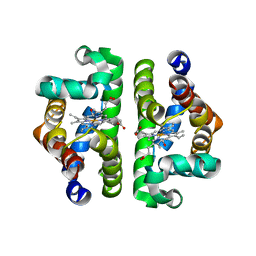 | | Crystal Structure of Scapharca inaequivalvis HbI, I114F Mutant Ligated to Carbon Monoxide. | | Descriptor: | CARBON MONOXIDE, Globin I - Ark Shell, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Knapp, J.E, Gibson, Q.H, Cushing, L, Royer Jr, W.E. | | Deposit date: | 2001-09-04 | | Release date: | 2001-12-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Restricting the ligand-linked heme movement in Scapharca dimeric hemoglobin reveals tight coupling between distal and proximal contributions to cooperativity.
Biochemistry, 40, 2001
|
|
4LCE
 
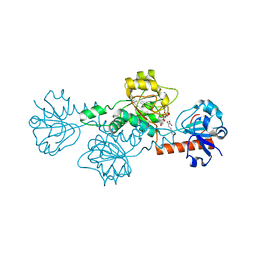 | | CtBP1 in complex with substrate MTOB | | Descriptor: | 4-(METHYLSULFANYL)-2-OXOBUTANOIC ACID, C-terminal-binding protein 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hilbert, B.J, Schiffer, C.A, Royer Jr, W.E. | | Deposit date: | 2013-06-21 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structures of human CtBP in complex with substrate MTOB reveal active site features useful for inhibitor design.
Febs Lett., 588, 2014
|
|
4LCJ
 
 | | CtBP2 in complex with substrate MTOB | | Descriptor: | 4-(METHYLSULFANYL)-2-OXOBUTANOIC ACID, C-terminal-binding protein 2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hilbert, B.J, Schiffer, C.A, Royer Jr, W.E. | | Deposit date: | 2013-06-21 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Crystal structures of human CtBP in complex with substrate MTOB reveal active site features useful for inhibitor design.
Febs Lett., 588, 2014
|
|
1NWI
 
 | |
1NWN
 
 | |
3QB7
 
 | | Interleukin-4 mutant RGA bound to cytokine receptor common gamma | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit gamma, ... | | Authors: | Bates, D.L, Junttila, I.S, Creusot, R.J, Moraga, I, Lupardus, P, Fathman, C.G, Paul, W.E, Garcia, K.C. | | Deposit date: | 2011-01-12 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.245 Å) | | Cite: | Redirecting cell-type specific cytokine responses with engineered interleukin-4 superkines.
Nat.Chem.Biol., 8, 2012
|
|
5T25
 
 | | Kinetic, Spectral and Structural Characterization of the Slow Binding Inhibitor Acetopyruvate with Dihydrodipicolinate Synthase from Escherichia coli. | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, LYSINE, SODIUM ION | | Authors: | Chooback, L, Thomas, L.M, Karsten, W.E, Fleming, C.D, Seabourn, P. | | Deposit date: | 2016-08-23 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | Kinetic, Spectral and Structural Characterization of the Slow Binding Inhibitor Acetopyruvate with Dihydrodipicolinate Synthase from Escherichia coli.
To Be Published
|
|
5T26
 
 | | Kinetic, Spectral and Structural Characterization of the Slow Binding Inhibitor Acetopyruvate with Dihydrodipicolinate Synthase from Escherichia coli. | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Chooback, L, Thomas, L.M, Karsten, W.E, Fleming, C.D, Seabourn, P. | | Deposit date: | 2016-08-23 | | Release date: | 2016-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic, Spectral and Structural Characterization of the Slow Binding Inhibitor Acetopyruvate with Dihydrodipicolinate Synthase from Escherichia coli.
To Be Published
|
|
1JZK
 
 | | Crystal Structure of Scapharca inaequivalvis HbI, I114F mutant (deoxy) | | Descriptor: | GLOBIN I - ARK SHELL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Knapp, J.E, Gibson, Q.H, Cushing, L, Royer Jr, W.E. | | Deposit date: | 2001-09-16 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Restricting the Ligand-Linked Heme Movement in Scapharca Dimeric Hemoglobin Reveals Tight Coupling between Distal and Proximal
Contributions to Cooperativity.
Biochemistry, 40, 2001
|
|
