6POE
 
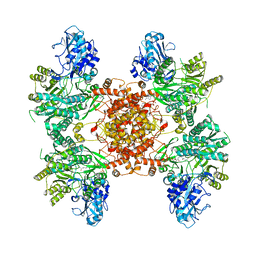 | | Structure of ACLY in complex with CoA | | Descriptor: | ATP-citrate synthase, COENZYME A | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2019-07-03 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for acetyl-CoA production by ATP-citrate lyase.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3S7H
 
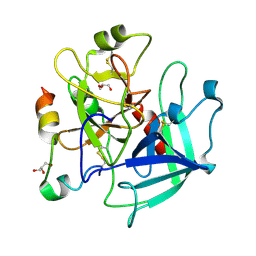 | | Structure of thrombin mutant Y225P in the E* form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Prothrombin | | Authors: | Niu, W, Chen, Z, Gandhi, P, Vogt, A, Pozzi, N, Pele, L.A, Zapata, F, Di Cera, E. | | Deposit date: | 2011-05-26 | | Release date: | 2011-07-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic and Kinetic Evidence of Allostery in a Trypsin-like Protease.
Biochemistry, 50, 2011
|
|
3S7K
 
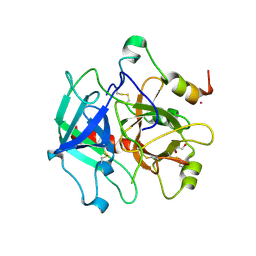 | | Structure of thrombin mutant Y225P in the E form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, POTASSIUM ION, Prothrombin | | Authors: | Niu, W, Chen, Z, Gandhi, P, Vogt, A, Pozzi, N, Pele, L.A, Zapata, F, Di Cera, E. | | Deposit date: | 2011-05-26 | | Release date: | 2011-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic and Kinetic Evidence of Allostery in a Trypsin-like Protease.
Biochemistry, 50, 2011
|
|
6UIA
 
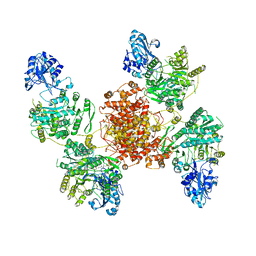 | |
7LJ9
 
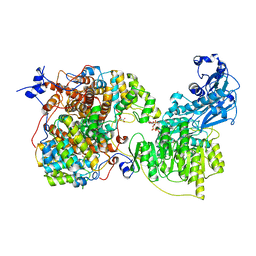 | |
7RIG
 
 | | Structure of ACLY-D1026A-substrates | | Descriptor: | (3S)-citryl-Coenzyme A, ADENOSINE-5'-DIPHOSPHATE, ATP-citrate synthase, ... | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2021-07-19 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Allosteric role of the citrate synthase homology domain of ATP citrate lyase.
Nat Commun, 14, 2023
|
|
7RKZ
 
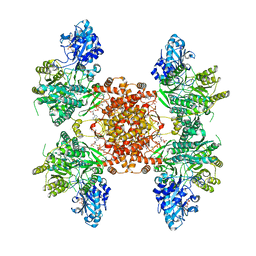 | | Structure of ACLY D1026A-substrates-asym-int | | Descriptor: | (3S)-citryl-Coenzyme A, ADENOSINE-5'-DIPHOSPHATE, ATP-citrate synthase, ... | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2021-07-22 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Allosteric role of the citrate synthase homology domain of ATP citrate lyase.
Nat Commun, 14, 2023
|
|
7RMP
 
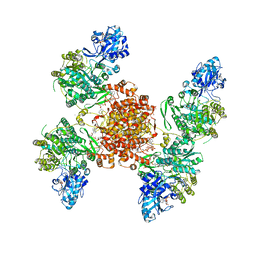 | | Structure of ACLY D1026A - substrates-asym | | Descriptor: | (3S)-citryl-Coenzyme A, ADENOSINE-5'-DIPHOSPHATE, ATP-citrate synthase, ... | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2021-07-28 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Allosteric role of the citrate synthase homology domain of ATP citrate lyase.
Nat Commun, 14, 2023
|
|
6GYH
 
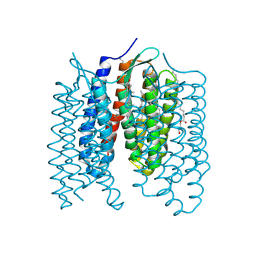 | | Crystal structure of the light-driven proton pump Coccomyxa subellipsoidea Rhodopsin CsR | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, Family A G protein-coupled receptor-like protein, ... | | Authors: | Szczepek, M, Schmidt, A, Scheerer, P. | | Deposit date: | 2018-06-29 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of a light-gated proton channel based on the crystal structure ofCoccomyxarhodopsin.
Sci.Signal., 12, 2019
|
|
6POF
 
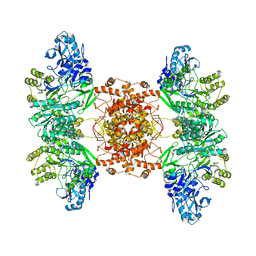 | | Structure of human ATP citrate lyase | | Descriptor: | ATP-citrate synthase | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2019-07-03 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Molecular basis for acetyl-CoA production by ATP-citrate lyase.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6UI9
 
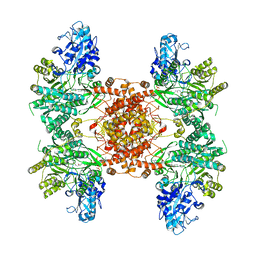 | |
6UUZ
 
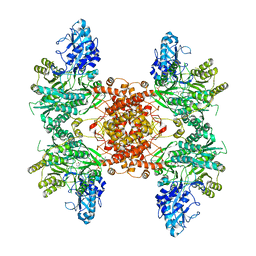 | |
6UV5
 
 | |
6UUW
 
 | | Structure of human ATP citrate lyase E599Q mutant in complex with Mg2+, citrate, ATP and CoA | | Descriptor: | (2S)-2-hydroxy-2-[2-oxo-2-(phosphonooxy)ethyl]butanedioic acid, ADENOSINE-5'-DIPHOSPHATE, ATP-citrate synthase, ... | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2019-11-01 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Molecular basis for acetyl-CoA production by ATP-citrate lyase
Nat.Struct.Mol.Biol., 27, 2020
|
|
8EZB
 
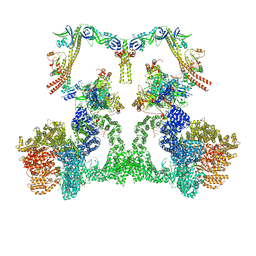 | | NHEJ Long-range complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (30-MER), DNA (31-MER), ... | | Authors: | Chen, S, He, Y. | | Deposit date: | 2022-10-31 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Cryo-EM visualization of DNA-PKcs structural intermediates in NHEJ.
Sci Adv, 9, 2023
|
|
8EZ9
 
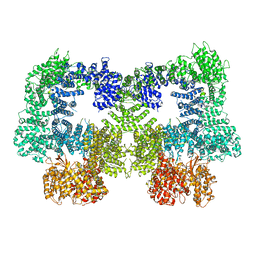 | | Dimeric complex of DNA-PKcs | | Descriptor: | DNA-dependent protein kinase catalytic subunit, unknown region of DNA-PKcs | | Authors: | Chen, S, He, Y. | | Deposit date: | 2022-10-31 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (5.67 Å) | | Cite: | Cryo-EM visualization of DNA-PKcs structural intermediates in NHEJ.
Sci Adv, 9, 2023
|
|
8EZA
 
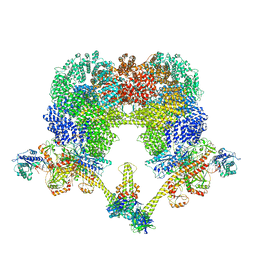 | | NHEJ Long-range complex with PAXX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (30-MER), DNA (31-MER), ... | | Authors: | Chen, S, He, Y. | | Deposit date: | 2022-10-31 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.39 Å) | | Cite: | Cryo-EM visualization of DNA-PKcs structural intermediates in NHEJ.
Sci Adv, 9, 2023
|
|
8G5C
 
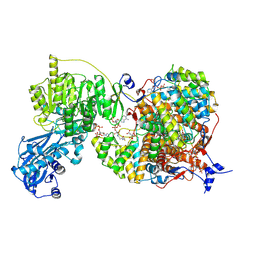 | |
8G1F
 
 | | Structure of ACLY-D1026A-products | | Descriptor: | (3S)-citryl-Coenzyme A, ACETYL COENZYME *A, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2023-02-02 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Allosteric role of the citrate synthase homology domain of ATP citrate lyase.
Nat Commun, 14, 2023
|
|
8G5D
 
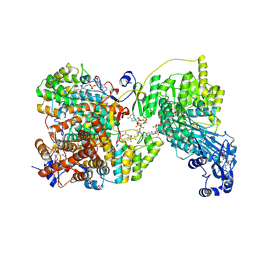 | |
8G1E
 
 | | Structure of ACLY-D1026A-products-asym | | Descriptor: | (3S)-citryl-Coenzyme A, ACETYL COENZYME *A, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2023-02-02 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Allosteric role of the citrate synthase homology domain of ATP citrate lyase.
Nat Commun, 14, 2023
|
|
8GHN
 
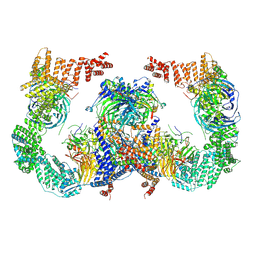 | |
8GHL
 
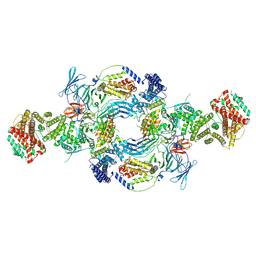 | | the Hir complex core | | Descriptor: | Histone transcription regulator 3, Protein HIR1, Protein HIR2 | | Authors: | Kim, H.J, Murakami, K. | | Deposit date: | 2023-03-10 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structure of the Hir histone chaperone complex.
Mol.Cell, 84, 2024
|
|
8GHA
 
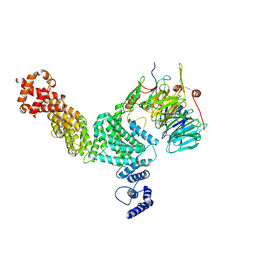 | | Hir3 Arm/Tail, Hir2 WD40, C-terminal Hpc2 | | Descriptor: | Histone promoter control protein 2, Histone transcription regulator 3, Protein HIR2 | | Authors: | Kim, H.J, Murakami, K. | | Deposit date: | 2023-03-09 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structure of the Hir histone chaperone complex.
Mol.Cell, 84, 2024
|
|
8GHM
 
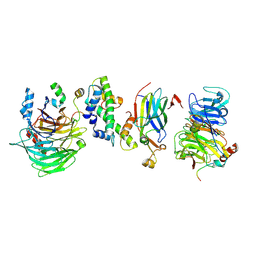 | | Hir1 WD40 domains and Asf1/H3/H4 | | Descriptor: | Histone H3, Histone H4, Histone chaperone, ... | | Authors: | Kim, H.J, Murakami, K. | | Deposit date: | 2023-03-10 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structure of the Hir histone chaperone complex.
Mol.Cell, 84, 2024
|
|
