2K36
 
 | | Structure ensemble Backbone and side-chain 1H, 13C, and 15N Chemical Shift Assignments, 1H-15N RDCs and 1H-1H nOe restraints for protein K7 from the Vaccinia virus | | Descriptor: | Protein K7 | | Authors: | Kalverda, A.P, Thompson, G.S, Vogel, A, Schr der, M, Bowie, A.G, Khan, A.R, Homans, S.W. | | Deposit date: | 2008-04-22 | | Release date: | 2008-10-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Poxvirus K7 protein adopts a Bcl-2 fold: biochemical mapping of its interactions with human DEAD box RNA helicase DDX3.
J.Mol.Biol., 385, 2009
|
|
8BRH
 
 | | Co-crystal structure of She4 with Myo4 peptide | | Descriptor: | KLLA0E16699p, Myo4 peptide (LYS-PHE-ILE-VAL-SER-HIS-TYR) | | Authors: | Arnese, R, Gudino, R, Meinhart, A, Clausen, T. | | Deposit date: | 2022-11-23 | | Release date: | 2024-06-05 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | UNC-45 assisted myosin folding depends on a conserved FX 3 HY motif implicated in Freeman Sheldon Syndrome.
Nat Commun, 15, 2024
|
|
8BRG
 
 | | Crystal structure of She4 | | Descriptor: | KLLA0E16699p | | Authors: | Gudino, R, Arnese, R, Meinhart, A, Clausen, T. | | Deposit date: | 2022-11-23 | | Release date: | 2024-06-05 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | UNC-45 assisted myosin folding depends on a conserved FX 3 HY motif implicated in Freeman Sheldon Syndrome.
Nat Commun, 15, 2024
|
|
5LHA
 
 | |
5LH9
 
 | |
7BII
 
 | | Crystal structure of Nematocida HUWE1 | | Descriptor: | E3 ubiquitin-protein ligase HUWE1 | | Authors: | Grabarczyk, D.B, Petrova, O.A, Meinhart, A, Kessler, D, Clausen, T. | | Deposit date: | 2021-01-12 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.037 Å) | | Cite: | HUWE1 employs a giant substrate-binding ring to feed and regulate its HECT E3 domain.
Nat.Chem.Biol., 17, 2021
|
|
6QDJ
 
 | | Molecular features of the UNC-45 chaperone critical for binding and folding muscle myosin | | Descriptor: | 1,4-BUTANEDIOL, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Meinhart, A, Clausen, T, Arnese, R. | | Deposit date: | 2019-01-02 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.884 Å) | | Cite: | Molecular features of the UNC-45 chaperone critical for binding and folding muscle myosin.
Nat Commun, 10, 2019
|
|
6QDL
 
 | |
6QDM
 
 | |
6QDK
 
 | |
6TAY
 
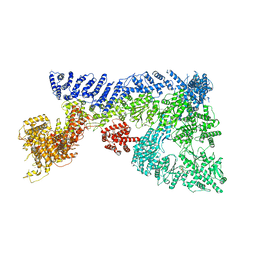 | | Mouse RNF213 mutant R4753K modeling the Moyamoya-disease-related Human variant R4810K | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNF213,E3 ubiquitin-protein ligase RNF213,E3 ubiquitin-protein ligase RNF213, ... | | Authors: | Ahel, J, Meinhart, A, Haselbach, D, Clausen, T. | | Deposit date: | 2019-10-31 | | Release date: | 2020-07-01 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Moyamoya disease factor RNF213 is a giant E3 ligase with a dynein-like core and a distinct ubiquitin-transfer mechanism.
Elife, 9, 2020
|
|
6TAX
 
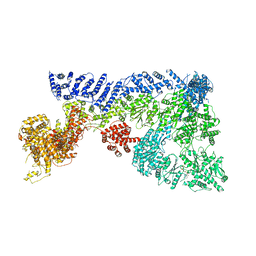 | | Mouse RNF213 wild type protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNF213,E3 ubiquitin-protein ligase RNF213,E3 ubiquitin-protein ligase RNF213, ... | | Authors: | Ahel, J, Meinhart, A, Haselbach, D, Clausen, T. | | Deposit date: | 2019-10-31 | | Release date: | 2020-07-01 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Moyamoya disease factor RNF213 is a giant E3 ligase with a dynein-like core and a distinct ubiquitin-transfer mechanism.
Elife, 9, 2020
|
|
2CBN
 
 | |
6TQ3
 
 | |
6TQ8
 
 | |
6TQ5
 
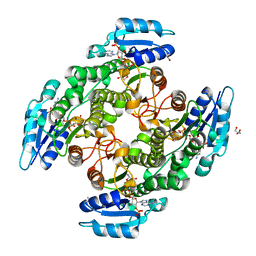 | | Alcohol dehydrogenase from Candida magnoliae DSMZ 70638 (ADHA): complex with NADP+ | | Descriptor: | Enzyme subunit, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Rovida, S, Aalbers, F.S, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2019-12-16 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Approaching boiling point stability of an alcohol dehydrogenase through computationally-guided enzyme engineering.
Elife, 9, 2020
|
|
6GIF
 
 | |
6GIG
 
 | |
2QUZ
 
 | |
2QZW
 
 | |
8G1Z
 
 | |
8G20
 
 | |
8PY3
 
 | | Crystal structure of human Sirt2 in complex with a 1,2,4-oxadiazole based inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-chloranyl-~{N}-[4-[5-[[(3~{S})-1-[(3-fluoranyl-2-methyl-phenyl)methyl]piperidin-3-yl]methyl]-1,2,4-oxadiazol-3-yl]phenyl]benzamide, ... | | Authors: | Friedrich, F, Colcerasa, A, Einsle, O, Jung, M. | | Deposit date: | 2023-07-24 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Activity Studies of 1,2,4-Oxadiazoles for the Inhibition of the NAD + -Dependent Lysine Deacylase Sirtuin 2.
J.Med.Chem., 67, 2024
|
|
7N03
 
 | | Crystal structure of MTH1 in complex with compound 31 | | Descriptor: | 4-anilino-6-(hexylamino)-N-methylquinoline-3-carboxamide, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Eron, S.J. | | Deposit date: | 2021-05-24 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Development of an AchillesTAG degradation system and its application to control CAR-T activity
Curr Res Chem Biol, 1, 2021
|
|
7N13
 
 | | Crystal structure of MTH1 in complex with compound 32 | | Descriptor: | 4-anilino-6-[4-(butylcarbamoyl)-3-fluorophenyl]-N-cyclopropyl-7-fluoroquinoline-3-carboxamide, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Eron, S.J. | | Deposit date: | 2021-05-26 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Development of an AchillesTAG degradation system and its application to control CAR-T activity
Curr Res Chem Biol, 1, 2021
|
|
