7UZE
 
 | | Erythrocyte ankyrin-1 complex class 2 local refinement of AQP1 (C4 symmetry applied) | | Descriptor: | Aquaporin-1, CHOLESTEROL | | Authors: | Vallese, F, Kim, K, Yen, L.Y, Johnston, J.D, Noble, A.J, Cali, T, Clarke, O.B. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-20 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Architecture of the human erythrocyte ankyrin-1 complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7V0U
 
 | | Local refinement of Band 3-II cytoplasmic domains, class 1 of erythrocyte ankyrin-1 complex | | Descriptor: | Band 3 anion transport protein | | Authors: | Vallese, F, Kim, K, Yen, L.Y, Johnston, J.D, Noble, A.J, Cali, T, Clarke, O.B. | | Deposit date: | 2022-05-11 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Architecture of the human erythrocyte ankyrin-1 complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7V0K
 
 | | Consensus refinement of human erythrocyte ankyrin-1 complex (Composite map) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ammonium transporter Rh type A, Ankyrin-1, ... | | Authors: | Vallese, F, Kim, K, Yen, L.Y, Johnston, J.D, Noble, A.J, Cali, T, Clarke, O.B. | | Deposit date: | 2022-05-10 | | Release date: | 2022-07-20 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Architecture of the human erythrocyte ankyrin-1 complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7V07
 
 | | Band 3-I-TM local refinement from erythrocyte ankyrin-1 complex consensus reconstruction | | Descriptor: | Band 3 anion transport protein, CHOLESTEROL, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Vallese, F, Kim, K, Yen, L.Y, Johnston, J.D, Noble, A.J, Cali, T, Clarke, O.B. | | Deposit date: | 2022-05-10 | | Release date: | 2022-07-20 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Architecture of the human erythrocyte ankyrin-1 complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7N9Z
 
 | | E. coli cytochrome bo3 in MSP nanodisc | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Vallese, F, Clarke, O.B. | | Deposit date: | 2021-06-19 | | Release date: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Cryo-EM structures of Escherichia coli cytochrome bo 3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4XFW
 
 | | Crystal structure of the monoclinic form of alpha-carbonic anhydrase from the human pathogen Helicobacter pylori | | Descriptor: | ACETIC ACID, Alpha-carbonic anhydrase, CHLORIDE ION, ... | | Authors: | Compostella, M.E, Vallese, F, Berto, P, Zanotti, G. | | Deposit date: | 2014-12-29 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.517 Å) | | Cite: | Structure of alpha-carbonic anhydrase from the human pathogen Helicobacter pylori.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4OW6
 
 | | Crystal structure of Diphtheria Toxin at acidic pH | | Descriptor: | Diphtheria toxin | | Authors: | Leka, O, Vallese, F, Pirazzini, M, Berto, P, Montecucco, C, Zanotti, G. | | Deposit date: | 2014-01-31 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Diphtheria toxin conformational switching at acidic pH.
Febs J., 281, 2014
|
|
3QQ5
 
 | | Crystal structure of the [FeFe]-hydrogenase maturation protein HydF | | Descriptor: | Small GTP-binding protein | | Authors: | Cendron, L, Berto, P, D'Adamo, S, Vallese, F, Govoni, C, Posewitz, M.C, Giacometti, G.M, Costantini, P, Zanotti, G. | | Deposit date: | 2011-02-15 | | Release date: | 2011-11-16 | | Last modified: | 2012-01-11 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal Structure of HydF Scaffold Protein Provides Insights into [FeFe]-Hydrogenase Maturation.
J.Biol.Chem., 286, 2011
|
|
5OQ0
 
 | |
5LJC
 
 | | Crystal structure of holo human CRBP1 | | Descriptor: | RETINOL, Retinol-binding protein 1, SODIUM ION | | Authors: | Zanotti, G, Vallese, F, Berni, R, Menozzi, I. | | Deposit date: | 2016-07-18 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.433 Å) | | Cite: | Structural and molecular determinants affecting the interaction of retinol with human CRBP1.
J. Struct. Biol., 197, 2017
|
|
5LJH
 
 | | Crystal structure of human apo CRBP1/K40L mutant | | Descriptor: | Retinol-binding protein 1, SODIUM ION | | Authors: | Zanotti, G, Vallese, F, Berni, R, Menozzi, I. | | Deposit date: | 2016-07-18 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural and molecular determinants affecting the interaction of retinol with human CRBP1.
J. Struct. Biol., 197, 2017
|
|
5LJD
 
 | | Crystal structure of holo human CRBP1/K40L mutant | | Descriptor: | RETINOL, Retinol-binding protein 1, SODIUM ION | | Authors: | Zanotti, G, Vallese, F, Berni, R, Menozzi, I. | | Deposit date: | 2016-07-18 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural and molecular determinants affecting the interaction of retinol with human CRBP1.
J. Struct. Biol., 197, 2017
|
|
5LJG
 
 | | Crystal structure of holo human CRBP1 | | Descriptor: | PALMITIC ACID, Retinol-binding protein 1 | | Authors: | Zanotti, G, Vallese, F, Berni, R, Menozzi, I. | | Deposit date: | 2016-07-18 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.146 Å) | | Cite: | Structural and molecular determinants affecting the interaction of retinol with human CRBP1.
J. Struct. Biol., 197, 2017
|
|
5LJK
 
 | | Crystal structure of human apo CRBP1 | | Descriptor: | Retinol-binding protein 1, SODIUM ION | | Authors: | Zanotti, G, Vallese, F, Berni, R, Menozzi, I. | | Deposit date: | 2016-07-18 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and molecular determinants affecting the interaction of retinol with human CRBP1.
J. Struct. Biol., 197, 2017
|
|
5LJE
 
 | | Crystal structure of holo human CRBP1/K40L,Q108L mutant | | Descriptor: | RETINOL, Retinol-binding protein 1, SODIUM ION | | Authors: | Zanotti, G, Vallese, F, Berni, R, Menozzi, I. | | Deposit date: | 2016-07-18 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and molecular determinants affecting the interaction of retinol with human CRBP1.
J. Struct. Biol., 197, 2017
|
|
5MII
 
 | |
5MIF
 
 | | Crystal structure of carboxyl esterase 2 (TmelEST2) from mycorrhizal fungus Tuber melanosporum | | Descriptor: | 'Carboxyl esterase 2, FRAGMENT OF TRITON X-100 | | Authors: | Zanotti, G, Vallese, F, Cavazzini, D, Ottonello, S. | | Deposit date: | 2016-11-28 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | A family of archaea-like carboxylesterases preferentially expressed in the symbiotic phase of the mycorrhizal fungus Tuber melanosporum.
Sci Rep, 7, 2017
|
|
7SIN
 
 | | Structure of negative allosteric modulator-bound inactive human calcium-sensing receptor | | Descriptor: | 2-chloro-6-[(2R)-2-hydroxy-3-{[2-methyl-1-(naphthalen-2-yl)propan-2-yl]amino}propoxy]benzonitrile, Isoform 1 of Extracellular calcium-sensing receptor | | Authors: | Park, J, Zuo, H, Frangaj, A, Fu, Z, Yen, L.Y, Zhang, Z, Mosyak, L, Slavkovich, V.N, Liu, J, Ray, K.M, Cao, B, Vallese, F, Geng, Y, Chen, S, Grassucci, R, Dandey, V.P, Tan, Y.Z, Eng, E, Lee, Y, Kloss, B, Liu, Z, Hendrickson, W.A, Potter, C.S, Carragher, B, Graziano, J, Conigrave, A.D, Frank, J, Clarke, O.B, Fan, Q.R. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Symmetric activation and modulation of the human calcium-sensing receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SIL
 
 | | Structure of positive allosteric modulator-bound active human calcium-sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-chlorophenyl)-N-[(1R)-1-(3-methoxyphenyl)ethyl]propan-1-amine, ... | | Authors: | Park, J, Zuo, H, Frangaj, A, Fu, Z, Yen, L.Y, Zhang, Z, Mosyak, L, Slavkovich, V.N, Liu, J, Ray, K.M, Cao, B, Vallese, F, Geng, Y, Chen, S, Grassucci, R, Dandey, V.P, Tan, Y.Z, Eng, E, Lee, Y, Kloss, B, Liu, Z, Hendrickson, W.A, Potter, C.S, Carragher, B, Graziano, J, Conigrave, A.D, Frank, J, Clarke, O.B, Fan, Q.R. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Symmetric activation and modulation of the human calcium-sensing receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SIM
 
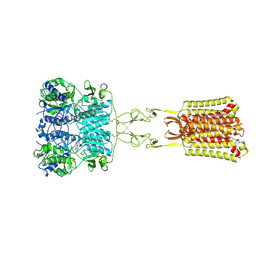 | | Structure of positive allosteric modulator-free active human calcium-sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Park, J, Zuo, H, Frangaj, A, Fu, Z, Yen, L.Y, Zhang, Z, Mosyak, L, Slavkovich, V.N, Liu, J, Ray, K.M, Cao, B, Vallese, F, Geng, Y, Chen, S, Grassucci, R, Dandey, V.P, Tan, Y.Z, Eng, E, Lee, Y, Kloss, B, Liu, Z, Hendrickson, W.A, Potter, C.S, Carragher, B, Graziano, J, Conigrave, A.D, Frank, J, Clarke, O.B, Fan, Q.R. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Symmetric activation and modulation of the human calcium-sensing receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5OK8
 
 | |
7OH1
 
 | |
7OH0
 
 | |
7CUW
 
 | | Ubiquinol Binding Site of Cytochrome bo3 from Escherichia coli | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | Deposit date: | 2020-08-25 | | Release date: | 2021-08-25 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CUQ
 
 | | 2.55-Angstrom Cryo-EM structure of Cytochrome bo3 from Escherichia coli in Native Membrane | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | Deposit date: | 2020-08-24 | | Release date: | 2021-08-25 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
