1OYZ
 
 | | X-RAY STRUCTURE OF YIBA_ECOLI NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ET31. | | Descriptor: | Hypothetical protein yibA | | Authors: | Kuzin, A, Semesi, A, Skarina, T, Savchenko, A, Arrowsmith, C, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-04-07 | | Release date: | 2003-08-12 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-RAY STRUCTURE OF YIBA_ECOLI NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ET31.
To be Published
|
|
3CFU
 
 | | Crystal structure of the yjhA protein from Bacillus subtilis. Northeast Structural Genomics Consortium target SR562 | | Descriptor: | Uncharacterized lipoprotein yjhA | | Authors: | Vorobiev, S.M, Chen, Y, Kuzin, A.P, Seetharaman, J, Forouhar, F, Zhao, L, Mao, L, Maglaqui, M, Xiao, R, Liu, J, Swapna, G, Huang, J.Y, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-03-04 | | Release date: | 2008-03-18 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the yjhA protein from Bacillus subtilis.
To be Published
|
|
2H4O
 
 | | X-ray Crystal Structure of Protein yonK from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR415 | | Descriptor: | YonK protein | | Authors: | Seetharaman, J, Sue, M, Forouhar, F, Ken, C, Bonnie, C, Ma, L, Xiao, R, Acton, T.B, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-05-24 | | Release date: | 2006-07-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the hypothetical protein from bacillus subtilis (yonk).
To be Published
|
|
3U02
 
 | | Crystal Structure of the tRNA modifier TiaS from Pyrococcus furiosus, Northeast Structural Genomics Consortium Target PfR225 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETIC ACID, CITRIC ACID, ... | | Authors: | Kuzin, A, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-28 | | Release date: | 2011-11-02 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Crystal Structure of the tRNA modifier TiaS from Pyrococcus furiosus, Northeast Structural Genomics Consortium Target PfR225
To be Published
|
|
1PG6
 
 | | X-Ray Crystal Structure of Protein SPYM3_0169 from Streptococcus pyogenes. Northeast Structural Genomics Consortium Target DR2. | | Descriptor: | CALCIUM ION, Hypothetical protein SpyM3_0169 | | Authors: | Kuzin, A, Lee, I, Edstrom, W, Xiao, R, Acton, T, Rost, B, Montelione, G, Hunt, J, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-05-27 | | Release date: | 2003-12-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of hypothetical protein SPYM3_0169 from Streptococcus pyogenes
To be Published, 2003
|
|
3U19
 
 | | CRYSTAL STRUCTURE OF ACYLENZYME INTERMEDIATE OF DE NOVO DESIGNED CYSTEINE ESTERASE ECH13, Northeast Structural Genomics Consortium Target OR51 | | Descriptor: | GLYCEROL, artificial protein OR51 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Kohan, E, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-29 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF ACYLENZYME INTERMEDIATE OF DE NOVO DESIGNED CYSTEINE
ESTERASE ECH13, Northeast Structural Genomics Consortium Target OR51
To be Published
|
|
3D0W
 
 | | Crystal structure of YflH protein from Bacillus subtilis. Northeast Structural Genomics Consortium target SR326 | | Descriptor: | YflH protein | | Authors: | Seetharaman, J, Kuzin, A.P, Neely, H, Forouhar, F, Min, S, Zhao, L, Fang, Y, Owens, L, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-05-02 | | Release date: | 2008-05-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of YflH protein from Bacillus subtilis.
To be Published
|
|
1PE0
 
 | |
3U9R
 
 | |
1PDV
 
 | |
1PDW
 
 | |
3U9T
 
 | |
1PJ4
 
 | | Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme in a pentary complex with natural substrate malate, ATP, Mn++, and allosteric activator fumarate. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-MALATE, FUMARIC ACID, ... | | Authors: | Tao, X, Yang, Z, Tong, L. | | Deposit date: | 2003-05-31 | | Release date: | 2003-09-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of substrate complexes of malic enzyme and insights into the catalytic mechanism.
Structure, 11, 2003
|
|
3U13
 
 | | Crystal Structure of de Novo design of cystein esterase ECH13 at the resolution 1.6A, Northeast Structural Genomics Consortium Target OR51 | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Kohan, E, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-29 | | Release date: | 2011-11-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
1PJ2
 
 | | Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme in a pentary complex with natural substrate malate, cofactor NADH, Mn++, and allosteric activator fumarate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FUMARIC ACID, ... | | Authors: | Tao, X, Yang, Z, Tong, L. | | Deposit date: | 2003-05-30 | | Release date: | 2003-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of substrate complexes of malic enzyme and insights into the catalytic mechanism.
Structure, 11, 2003
|
|
3UAK
 
 | | Crystal Structure of De Novo designed cysteine esterase ECH14, Northeast Structural Genomics Consortium Target OR54 | | Descriptor: | De Novo designed cysteine esterase ECH14 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Richter, F, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-10-21 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.232 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
1PJ3
 
 | | Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme in a pentary complex with natural substrate pyruvate, cofactor NAD+, Mn++, and allosteric activator fumarate. | | Descriptor: | FUMARIC ACID, MANGANESE (II) ION, NAD-dependent malic enzyme, ... | | Authors: | Tao, X, Yang, Z, Tong, L. | | Deposit date: | 2003-05-30 | | Release date: | 2003-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of substrate complexes of malic enzyme and insights into the catalytic mechanism.
Structure, 11, 2003
|
|
3U07
 
 | | Crystal Structure of the VPA0106 protein from Vibrio parahaemolyticus. Northeast Structural Genomics Consortium Target VpR106. | | Descriptor: | uncharacterized protein VPA0106 | | Authors: | Vorobiev, S, Neely, H, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-28 | | Release date: | 2011-10-12 | | Method: | X-RAY DIFFRACTION (2.083 Å) | | Cite: | Crystal Structure of the VPA0106 protein from Vibrio parahaemolyticus.
To be Published
|
|
3U1O
 
 | | THREE DIMENSIONAL STRUCTURE OF DE NOVO DESIGNED CYSTEINE ESTERASE ECH19, Northeast Structural Genomics Consortium Target OR49 | | Descriptor: | De Novo design cysteine esterase ECH19, SODIUM ION, SULFATE ION | | Authors: | Kuzin, A, Su, M, Lew, S, Forouhar, F, Seetharaman, J, Daya, P, Xiao, R, Ciccosanti, C, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-30 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
3U1V
 
 | | X-ray Structure of De Novo design cysteine esterase FR29, Northeast Structural Genomics Consortium Target OR52 | | Descriptor: | De Novo design cysteine esterase FR29 | | Authors: | Kuzin, A, Su, M, Vorobiev, S.M, Seetharaman, J, Patel, D, Xiao, R, Ciccosanti, C, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
3U4T
 
 | | Crystal Structure of the C-terminal part of the TPR repeat-containing protein Q11TI6_CYTH3 from Cytophaga hutchinsonii. Northeast Structural Genomics Consortium Target ChR11B. | | Descriptor: | TPR repeat-containing protein | | Authors: | Vorobiev, S, Neely, H, Chen, Y, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Maglaqui, M, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-10-10 | | Release date: | 2011-10-19 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | Crystal Structure of the C-terminal part of the TPR repeat-containing protein Q11TI6_CYTH3 from Cytophaga hutchinsonii.
To be Published
|
|
3D3N
 
 | | Crystal structure of lipase/esterase (lp_2923) from Lactobacillus plantarum. Northeast Structural Genomics Consortium target LpR108 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, Putative lipase/esterase | | Authors: | Forouhar, F, Su, M, Seetharaman, J, Mao, L, Janjua, H, Xiao, R, Ciccosanti, C, Maglaqui, M, Foote, E.L, Zhao, L, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-05-12 | | Release date: | 2008-06-10 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of lipase/esterase (lp_2923) from Lactobacillus plantarum. Northeast Structural Genomics Consortium target LpR108
To be Published
|
|
3U9S
 
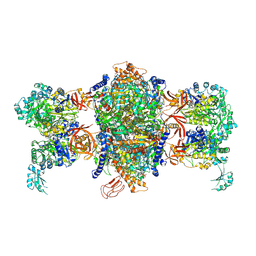 | | Crystal structure of P. aeruginosa 3-methylcrotonyl-CoA carboxylase (MCC) 750 kD holoenzyme, CoA complex | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, COENZYME A, Methylcrotonyl-CoA carboxylase, ... | | Authors: | Huang, C.S, Tong, L. | | Deposit date: | 2011-10-19 | | Release date: | 2011-12-14 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | An unanticipated architecture of the 750-kDa {alpha}6{beta}6 holoenzyme of 3-methylcrotonyl-CoA carboxylase
Nature, 481, 2012
|
|
3CEU
 
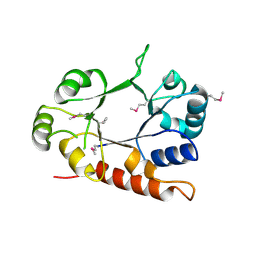 | | Crystal structure of thiamine phosphate pyrophosphorylase (BT_0647) from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium target BtR268 | | Descriptor: | Thiamine phosphate pyrophosphorylase | | Authors: | Forouhar, F, Chen, Y, Seetharaman, J, Janjua, H, Mao, L, Xiao, R, Foote, E.L, Maglaqui, M, Ciccosanti, C, Zhao, L, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-02-29 | | Release date: | 2008-03-25 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of thiamine phosphate pyrophosphorylase (BT_0647) from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium target BtR268.
To be Published
|
|
1PJL
 
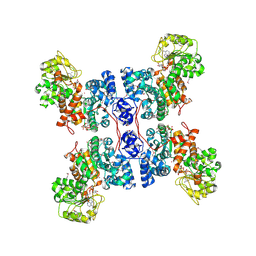 | | Crystal structure of human m-NAD-ME in ternary complex with NAD and Lu3+ | | Descriptor: | LUTETIUM (III) ION, NAD-dependent malic enzyme, mitochondrial, ... | | Authors: | Yang, Z, Batra, R, Floyd, D.L, Hung, H.-C, Chang, G.-G, Tong, L. | | Deposit date: | 2003-06-03 | | Release date: | 2003-06-17 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Potent and competitive inhibition of malic enzymes by lanthanide ions
Biochem.Biophys.Res.Commun., 274, 2000
|
|
