1LFO
 
 | | LIVER FATTY ACID BINDING PROTEIN-OLEATE COMPLEX | | Descriptor: | BUTENOIC ACID, LIVER FATTY ACID BINDING PROTEIN, OLEIC ACID, ... | | Authors: | Thompson, J, Winter, N, Terwey, D, Bratt, J, Banaszak, L. | | Deposit date: | 1996-12-09 | | Release date: | 1997-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the liver fatty acid-binding protein. A complex with two bound oleates.
J.Biol.Chem., 272, 1997
|
|
3ZD0
 
 | | The Solution Structure of Monomeric Hepatitis C Virus p7 Yields Potent Inhibitors of Virion Release | | Descriptor: | P7 PROTEIN | | Authors: | Foster, T.L, Sthompson, G, Kalverda, A.P, Kankanala, J, Thompson, J, Barker, A.M, Clarke, D, Noerenberg, M, Pearson, A.R, Rowlands, D.J, Homans, S.W, Harris, M, Foster, R, Griffin, S.D.C. | | Deposit date: | 2012-11-23 | | Release date: | 2013-09-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure-Guided Design Affirms Inhibitors of Hepatitis C Virus P7 as a Viable Class of Antivirals Targeting Virion Release
Hepatology, 59, 2014
|
|
3SQF
 
 | | Crystal structure of monomeric M-PMV retroviral protease | | Descriptor: | Protease | | Authors: | Jaskolski, M, Kazmierczyk, M, Gilski, M, Krzywda, S, Pichova, I, Zabranska, H, Khatib, F, DiMaio, F, Cooper, S, Thompson, J, Popovic, Z, Baker, D, Group, Foldit Contenders | | Deposit date: | 2011-07-05 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6324 Å) | | Cite: | Crystal structure of a monomeric retroviral protease solved by protein folding game players.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1DC9
 
 | | PROPERTIES AND CRYSTAL STRUCTURE OF A BETA-BARREL FOLDING MUTANT, V60N INTESTINAL FATTY ACID BINDING PROTEIN (IFABP) | | Descriptor: | INTESTINAL FATTY ACID BINDING PROTEIN | | Authors: | Ropson, I.J, Yowler, B.C, Dalessio, P.M, Banaszak, L, Thompson, J. | | Deposit date: | 1999-11-04 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Properties and crystal structure of a beta-barrel folding mutant.
Biophys.J., 78, 2000
|
|
4IPN
 
 | | The complex structure of 6-phospho-beta-glucosidase BglA-2 with thiocellobiose-6P from Streptococcus pneumoniae | | Descriptor: | 6-O-phosphono-alpha-L-idopyranose-(1-4)-4-thio-beta-D-glucopyranose, 6-phospho-beta-glucosidase | | Authors: | Yu, W.L, Jiang, Y.L, Andreas, P, Cheng, W, Bai, X.H, Ren, Y.M, Thompsonn, J, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2013-01-10 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Structural insights into the substrate specificity of a 6-phospho-&[beta]-glucosidase BglA-2 from Streptococcus pneumoniae TIGR4
J.Biol.Chem., 288, 2013
|
|
4IPL
 
 | | The crystal structure of 6-phospho-beta-glucosidase BglA-2 from Streptococcus pneumoniae | | Descriptor: | 6-phospho-beta-glucosidase, GLYCEROL | | Authors: | Yu, W.L, Jiang, Y.L, Andreas, P, Cheng, W, Bai, X.H, Ren, Y.M, Thompsonn, J, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2013-01-10 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Structural insights into the substrate specificity of a 6-phospho-&[beta]-glucosidase BglA-2 from Streptococcus pneumoniae TIGR4
J.Biol.Chem., 288, 2013
|
|
6S1U
 
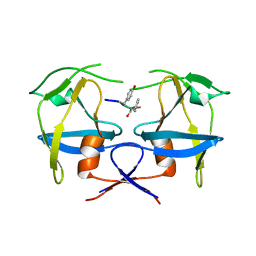 | | Crystal structure of dimeric M-PMV protease C7A/D26N/C106A mutant in complex with inhibitor | | Descriptor: | Gag-Pro-Pol polyprotein, PRO-0A1-VAL-PSA-ALA-MET-THR | | Authors: | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | Deposit date: | 2019-06-19 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparison of a retroviral protease in monomeric and dimeric states.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6S1W
 
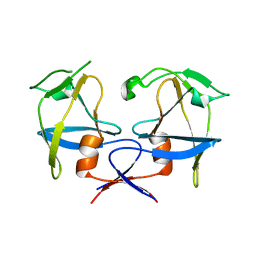 | | Crystal structure of dimeric M-PMV protease D26N mutant | | Descriptor: | Gag-Pro-Pol polyprotein | | Authors: | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | Deposit date: | 2019-06-19 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Comparison of a retroviral protease in monomeric and dimeric states.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6S1V
 
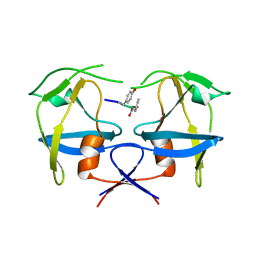 | | Crystal structure of dimeric M-PMV protease D26N mutant in complex with inhibitor | | Descriptor: | Gag-Pro-Pol polyprotein, PRO-0A1-VAL-PSA-ALA-MET-THR | | Authors: | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | Deposit date: | 2019-06-19 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Comparison of a retroviral protease in monomeric and dimeric states.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
7BGU
 
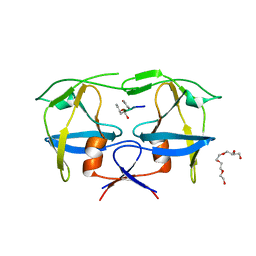 | | Mason-Pfizer Monkey Virus Protease mutant C7A/D26N/C106A in complex with peptidomimetic inhibitor | | Descriptor: | Gag-Pro-Pol polyprotein, PENTAETHYLENE GLYCOL, peptidomimetic inhibitor | | Authors: | Wosicki, S, Gilski, M, Kazmierczyk, M, Jaskolski, M, Zabranska, H, Pichova, I. | | Deposit date: | 2021-01-08 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.433 Å) | | Cite: | Crystal structures of inhibitor complexes of M-PMV protease with visible flap loops.
Protein Sci., 30, 2021
|
|
7BGT
 
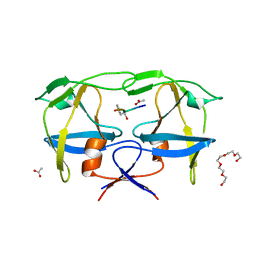 | | Mason-Pfizer Monkey Virus Protease mutant C7A/D26N/C106A in complex with peptidomimetic inhibitor | | Descriptor: | ACETATE ION, Gag-Pro-Pol polyprotein, PENTAETHYLENE GLYCOL, ... | | Authors: | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | Deposit date: | 2021-01-08 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of inhibitor complexes of M-PMV protease with visible flap loops.
Protein Sci., 30, 2021
|
|
1ALY
 
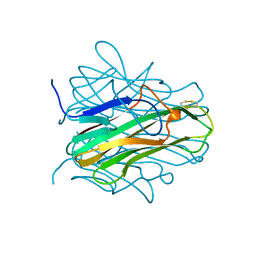 | |
1MMS
 
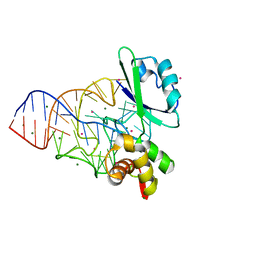 | | Crystal structure of the ribosomal PROTEIN L11-RNA complex | | Descriptor: | 23S RIBOSOMAL RNA, CADMIUM ION, MAGNESIUM ION, ... | | Authors: | Wimberly, B.T, Guymon, R, Mccutcheon, J.P, White, S.W, Ramakrishnan, V. | | Deposit date: | 1999-04-14 | | Release date: | 2000-04-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | A detailed view of a ribosomal active site: the structure of the L11-RNA complex.
Cell(Cambridge,Mass.), 97, 1999
|
|
3DBN
 
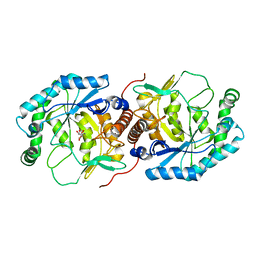 | | Crystal structure of the Streptoccocus suis serotype2 D-mannonate dehydratase in complex with its substrate | | Descriptor: | D-MANNONIC ACID, MANGANESE (II) ION, Mannonate dehydratase | | Authors: | Peng, H, Zhang, Q, Gao, F, Gao, G.F. | | Deposit date: | 2008-06-02 | | Release date: | 2009-06-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of Streptococcus suis mannonate dehydratase (ManD) and its complex with substrate: genetic and biochemical evidence for a catalytic mechanism
J.Bacteriol., 191, 2009
|
|
7RRO
 
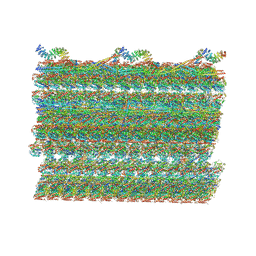 | | Structure of the 48-nm repeat doublet microtubule from bovine tracheal cilia | | Descriptor: | Armadillo repeat containing 4, Chromosome 3 C1orf194 homolog, Cilia and flagella associated protein 161, ... | | Authors: | Gui, M, Anderson, J.R, Botsch, J.J, Meleppattu, S, Singh, S.K, Zhang, Q, Brown, A. | | Deposit date: | 2021-08-10 | | Release date: | 2021-10-27 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | De novo identification of mammalian ciliary motility proteins using cryo-EM.
Cell, 184, 2021
|
|
3QDD
 
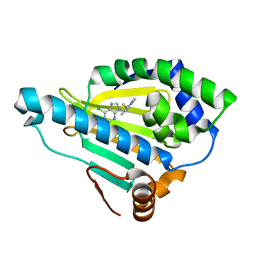 | | HSP90A N-terminal domain in complex with BIIB021 | | Descriptor: | 6-chloro-9-[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]-9H-purin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Arndt, J.W, Biamonte, M.A. | | Deposit date: | 2011-01-18 | | Release date: | 2012-07-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | EC144 Is a Potent Inhibitor of the Heat Shock Protein 90.
J.Med.Chem., 55, 2012
|
|
3FVM
 
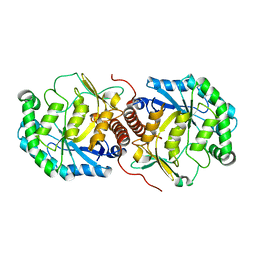 | | Crystal structure of Steptococcus suis mannonate dehydratase with metal Mn++ | | Descriptor: | MANGANESE (II) ION, Mannonate dehydratase | | Authors: | Peng, H, Zhang, Q.J, Gao, F, Liu, Y, Gao, F.G. | | Deposit date: | 2009-01-16 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of Streptococcus suis mannonate dehydratase (ManD) and its complex with substrate: genetic and biochemical evidence for a catalytic mechanism
J.Bacteriol., 191, 2009
|
|
1UP7
 
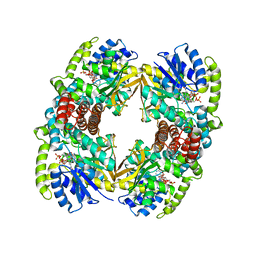 | | Structure of the 6-phospho-beta glucosidase from Thermotoga maritima at 2.4 Angstrom resolution in the tetragonal form with NAD and glucose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, 6-PHOSPHO-BETA-GLUCOSIDASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Varrot, A, Yip, V.L, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-09-29 | | Release date: | 2004-11-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nad+ and Metal-Ion Dependent Hydrolysis by Family 4 Glycosidases: Structural Insight Into Specificity for Phospho-Beta-D-Glucosides
J.Mol.Biol., 346, 2005
|
|
1UP6
 
 | | Structure of the 6-phospho-beta glucosidase from Thermotoga maritima at 2.55 Angstrom resolution in the tetragonal form with manganese, NAD+ and glucose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, 6-PHOSPHO-BETA-GLUCOSIDASE, MANGANESE (II) ION, ... | | Authors: | Varrot, A, Yip, V.L, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-09-29 | | Release date: | 2004-08-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | An Unusual Mechanism of Glycoside Hydrolysis Involving Redox and Elimination Steps by a Family 4 Beta-Glycosidase from Thermotoga Maritima.
J.Am.Chem.Soc., 126, 2004
|
|
1UP4
 
 | |
1U8X
 
 | | CRYSTAL STRUCTURE OF GLVA FROM BACILLUS SUBTILIS, A METAL-REQUIRING, NAD-DEPENDENT 6-PHOSPHO-ALPHA-GLUCOSIDASE | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, MANGANESE (II) ION, Maltose-6'-phosphate glucosidase, ... | | Authors: | Rajan, S.S, Yang, X, Collart, F, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-09 | | Release date: | 2004-08-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Novel Catalytic Mechanism of Glycoside Hydrolysis Based on the Structure of an NAD(+)/Mn(2+)-Dependent Phospho-alpha-Glucosidase from Bacillus subtilis.
STRUCTURE, 12, 2004
|
|
3O03
 
 | | Quaternary complex structure of gluconate 5-dehydrogenase from streptococcus suis type 2 | | Descriptor: | CALCIUM ION, D-gluconic acid, Dehydrogenase with different specificities, ... | | Authors: | Peng, H, Gao, F, Zhang, Q, Liu, Y, Gao, G.F. | | Deposit date: | 2010-07-18 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insight Into the Catalytic Mechanism of Gluconate 5-Dehydrogenase from Streptococcus Suis: Crystal Structures of the Substrate-Free and Quaternary Complex Enzymes.
Protein Sci., 18, 2009
|
|
3GC9
 
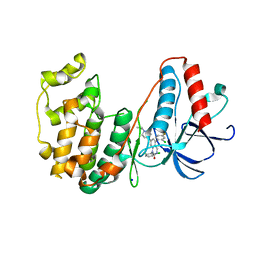 | | The structure of p38beta C119S, C162S in complex with a dihydroquinazolinone inhibitor | | Descriptor: | 5-(2-chloro-4-fluorophenyl)-1-(2,6-dichlorophenyl)-7-[1-(1-methylethyl)piperidin-4-yl]-3,4-dihydroquinazolin-2(1H)-one, Mitogen-activated protein kinase 11, SODIUM ION, ... | | Authors: | Scapin, G, Patel, S.B. | | Deposit date: | 2009-02-21 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The three-dimensional structure of MAP kinase p38beta: different features of the ATP-binding site in p38beta compared with p38alpha.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3GC8
 
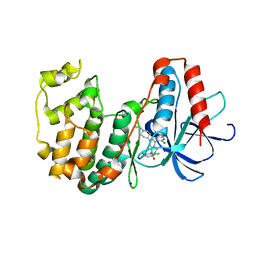 | | The structure of p38beta C162S in complex with a dihydroquinazolinone | | Descriptor: | 5-(2-chloro-4-fluorophenyl)-1-(2,6-dichlorophenyl)-7-[1-(1-methylethyl)piperidin-4-yl]-3,4-dihydroquinazolin-2(1H)-one, Mitogen-activated protein kinase 11, NICKEL (II) ION, ... | | Authors: | Scapin, G, Patel, S.B. | | Deposit date: | 2009-02-21 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure of MAP kinase p38beta: different features of the ATP-binding site in p38beta compared with p38alpha.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3GC7
 
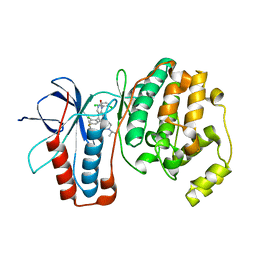 | | The structure of p38alpha in complex with a dihydroquinazolinone | | Descriptor: | 5-(2-chloro-4-fluorophenyl)-1-(2,6-dichlorophenyl)-7-[1-(1-methylethyl)piperidin-4-yl]-3,4-dihydroquinazolin-2(1H)-one, Mitogen-activated protein kinase 14 | | Authors: | Scapin, G, Patel, S.B. | | Deposit date: | 2009-02-21 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The three-dimensional structure of MAP kinase p38beta: different features of the ATP-binding site in p38beta compared with p38alpha.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
