3AA4
 
 | | A52V E.coli RNase HI | | Descriptor: | Ribonuclease HI | | Authors: | Takano, K. | | Deposit date: | 2009-11-11 | | Release date: | 2010-10-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Protein core adaptability: crystal structures of the cavity-filling variants of Escherichia coli RNase HI
Protein Pept.Lett., 17, 2010
|
|
1WMA
 
 | | Crystal structure of human CBR1 in complex with Hydroxy-PP | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 3-(4-AMINO-1-TERT-BUTYL-1H-PYRAZOLO[3,4-D]PYRIMIDIN-3-YL)PHENOL, ... | | Authors: | Rauh, D, Bateman, R, Shokat, K.M. | | Deposit date: | 2004-07-06 | | Release date: | 2005-04-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | An unbiased cell morphology-based screen for new, biologically active small molecules
Plos Biol., 3, 2005
|
|
7YV1
 
 | | Human K-Ras G12D (GDP-bound) in complex with cyclic peptide inhibitor LUNA18 and KA30L Fab | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, KA30L Fab H-chain, ... | | Authors: | Irie, M, Fukami, T.A, Matsuo, A, Saka, K, Nishimura, M, Saito, H, Torizawa, T, Tanada, M, Ohta, A. | | Deposit date: | 2022-08-18 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.454 Å) | | Cite: | Validation of a New Methodology to Create Oral Drugs beyond the Rule of 5 for Intracellular Tough Targets.
J.Am.Chem.Soc., 145, 2023
|
|
1AHL
 
 | | ANTHOPLEURIN-A,NMR, 20 STRUCTURES | | Descriptor: | ANTHOPLEURIN-A | | Authors: | Pallaghy, P.K, Scanlon, M.J, Monks, S.A, Norton, R.S. | | Deposit date: | 1994-10-28 | | Release date: | 1995-11-14 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure in solution of the polypeptide cardiac stimulant anthopleurin-A.
Biochemistry, 34, 1995
|
|
7YUZ
 
 | | Human K-Ras G12D (GDP-bound) in complex with cyclic peptide inhibitor AP8784 | | Descriptor: | AP8784, GUANOSINE-5'-DIPHOSPHATE, IODIDE ION, ... | | Authors: | Irie, M, Fukami, T.A, Tanada, M, Ohta, A, Torizawa, T. | | Deposit date: | 2022-08-18 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.878 Å) | | Cite: | Validation of a New Methodology to Create Oral Drugs beyond the Rule of 5 for Intracellular Tough Targets.
J.Am.Chem.Soc., 145, 2023
|
|
3W9Z
 
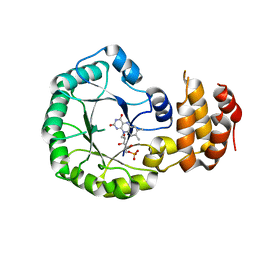 | | Crystal structure of DusC | | Descriptor: | FLAVIN MONONUCLEOTIDE, tRNA-dihydrouridine synthase C | | Authors: | Chen, M, Yu, J, Tanaka, Y, Tanaka, I, Yao, M. | | Deposit date: | 2013-04-19 | | Release date: | 2013-07-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of dihydrouridine synthase C (DusC) from Escherichia coli
Acta Crystallogr.,Sect.F, 69, 2013
|
|
6E7D
 
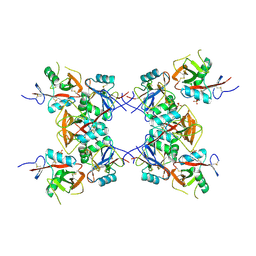 | | Structure of the inhibitory NKR-P1B receptor bound to the host-encoded ligand, Clr-b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain family 2 member D, Killer cell lectin-like receptor subfamily B member 1B allele B, ... | | Authors: | Balaji, G.R, Rossjohn, J, Berry, R. | | Deposit date: | 2018-07-26 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Recognition of host Clr-b by the inhibitory NKR-P1B receptor provides a basis for missing-self recognition.
Nat Commun, 9, 2018
|
|
6AI6
 
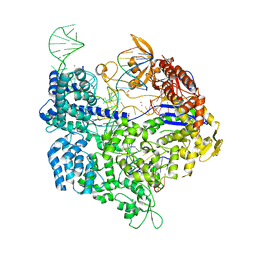 | | Crystal structure of SpCas9-NG | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9/Csn1, DNA (28-MER), ... | | Authors: | Nishimasu, H, Hirano, S, Ishitani, R, Nureki, O. | | Deposit date: | 2018-08-21 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Engineered CRISPR-Cas9 nuclease with expanded targeting space
Science, 361, 2018
|
|
5TZN
 
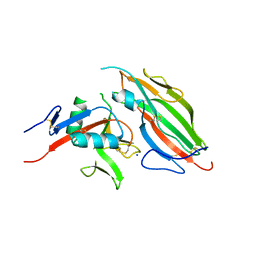 | | Structure of the viral immunoevasin m12 (Smith) bound to the natural killer cell receptor NKR-P1B (B6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein family protein m12, ... | | Authors: | Berry, R, Rossjohn, J. | | Deposit date: | 2016-11-22 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Viral Immunoevasin Controls Innate Immunity by Targeting the Prototypical Natural Killer Cell Receptor Family.
Cell, 169, 2017
|
|
6KS2
 
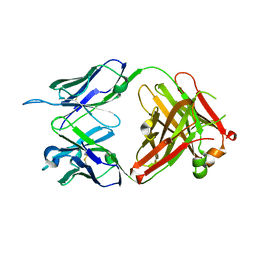 | | Structure of anti-Ghrelin receptor antibody | | Descriptor: | Fab7881 Heavy Chain (FabH), Fab7881 Light Chain (FabL) | | Authors: | Shiimura, Y, Horita, S, Asada, H, Hirata, K, Iwata, S, Kojima, M. | | Deposit date: | 2019-08-23 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.753 Å) | | Cite: | Structure of an antagonist-bound ghrelin receptor reveals possible ghrelin recognition mode.
Nat Commun, 11, 2020
|
|
6KO5
 
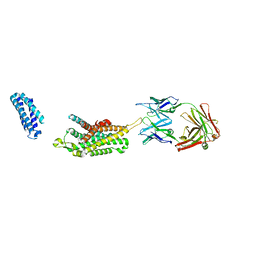 | | Complex structure of Ghrelin receptor with Fab | | Descriptor: | 6-(4-bromanyl-2-fluoranyl-phenoxy)-2-methyl-3-[[(3~{S})-1-propan-2-ylpiperidin-3-yl]methyl]pyrido[3,2-d]pyrimidin-4-one, Chimera of Soluble cytochrome b562 and Growth hormone secretagogue receptor type 1, Fab7881 Heavy Chain, ... | | Authors: | Shiimura, Y, Horita, S, Asada, H, Hirata, K, Iwata, S, Kojima, M. | | Deposit date: | 2019-08-08 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of an antagonist-bound ghrelin receptor reveals possible ghrelin recognition mode.
Nat Commun, 11, 2020
|
|
5ZG3
 
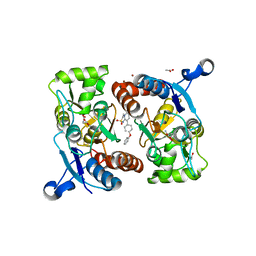 | | Crystal structure of the GluA2o LBD in complex with glutamate and TAK-137 | | Descriptor: | 9-(4-phenoxyphenyl)-3,4-dihydro-2H-2lambda~6~-pyrido[2,1-c][1,2,4]thiadiazine-2,2-dione, ACETATE ION, GLUTAMIC ACID, ... | | Authors: | Sogabe, S, Igaki, S, Hirokawa, A, Zama, Y, Lane, W, Snell, G. | | Deposit date: | 2018-03-07 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | TAK-137, an AMPA-R potentiator with little agonistic effect, has a wide therapeutic window.
Neuropsychopharmacology, 44, 2019
|
|
5ZG1
 
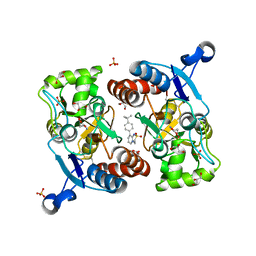 | | Crystal structure of the GluA2o LBD in complex with glutamate and Compound-2 | | Descriptor: | 9-(4-~{tert}-butylphenyl)-3,4-dihydropyrido[2,1-c][1,2,4]thiadiazine 2,2-dioxide, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Sogabe, S, Igaki, S, Hirokawa, A, Zama, Y, Lane, W, Snell, G. | | Deposit date: | 2018-03-07 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | TAK-137, an AMPA-R potentiator with little agonistic effect, has a wide therapeutic window.
Neuropsychopharmacology, 44, 2019
|
|
5ZG2
 
 | | Crystal structure of the GluA2o LBD in complex with ZK200775 and Compound-2 | | Descriptor: | 9-(4-~{tert}-butylphenyl)-3,4-dihydropyrido[2,1-c][1,2,4]thiadiazine 2,2-dioxide, ACETATE ION, GLYCEROL, ... | | Authors: | Sogabe, S, Igaki, S, Hirokawa, A, Zama, Y, Lane, W, Snell, G. | | Deposit date: | 2018-03-07 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | TAK-137, an AMPA-R potentiator with little agonistic effect, has a wide therapeutic window.
Neuropsychopharmacology, 44, 2019
|
|
5ZG0
 
 | | Crystal structure of the GluA2o LBD in complex with glutamate and Compound-1 | | Descriptor: | 9-{4-[(propan-2-yl)oxy]phenyl}-3,4-dihydro-2H-2lambda~6~-pyrido[2,1-c][1,2,4]thiadiazine-2,2-dione, ACETATE ION, GLUTAMIC ACID, ... | | Authors: | Sogabe, S, Igaki, S, Hirokawa, A, Zama, Y, Lane, W, Snell, G. | | Deposit date: | 2018-03-07 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | TAK-137, an AMPA-R potentiator with little agonistic effect, has a wide therapeutic window.
Neuropsychopharmacology, 44, 2019
|
|
2YV0
 
 | | Structural and Thermodynamic Analyses of E. coli ribonuclease HI Variant with Quintuple Thermostabilizing Mutations | | Descriptor: | Ribonuclease HI | | Authors: | Haruki, M, Motegi, T, Tadokoro, T, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-04-06 | | Release date: | 2008-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and thermodynamic analyses of Escherichia coli RNase HI variant with quintuple thermostabilizing mutations.
Febs J., 274, 2007
|
|
5ZFU
 
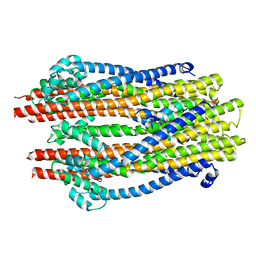 | | Structure of the ExbB/ExbD hexameric complex (ExbB6ExbD3TM) | | Descriptor: | 22-mer peptide from Biopolymer transport protein ExbD, Biopolymer transport protein ExbB | | Authors: | Yonekura, K, Yamashita, Y, Matsuoka, R, Maki-Yonekura, S. | | Deposit date: | 2018-03-07 | | Release date: | 2018-05-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Hexameric and pentameric complexes of the ExbBD energizer in the Ton system.
Elife, 7, 2018
|
|
5ZFP
 
 | |
5ZFV
 
 | | Structure of the ExbB/ExbD pentameric complex (ExbB5ExbD1TM) | | Descriptor: | 22-mer peptide from Biopolymer transport protein ExbD, Biopolymer transport protein ExbB | | Authors: | Yonekura, K, Yamashita, Y, Matsuoka, R, Maki-Yonekura, S. | | Deposit date: | 2018-03-07 | | Release date: | 2018-05-09 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Hexameric and pentameric complexes of the ExbBD energizer in the Ton system.
Elife, 7, 2018
|
|
7YL6
 
 | | Cell surface protein YwfG protein complexed with alpha-1,2-mannobiose | | Descriptor: | CALCIUM ION, GRAM_POS_ANCHORING domain-containing protein, SULFATE ION, ... | | Authors: | Tsuchiya, W, Fujimoto, Z, Suzuki, C. | | Deposit date: | 2022-07-25 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Cell-surface protein YwfG of Lactococcus lactis binds to alpha-1,2-linked mannose.
Plos One, 18, 2023
|
|
7YL5
 
 | | Cell surface protein YwfG protein complexed with mannose | | Descriptor: | CALCIUM ION, GRAM_POS_ANCHORING domain-containing protein, SULFATE ION, ... | | Authors: | Tsuchiya, W, Fujimoto, Z, Suzuki, C. | | Deposit date: | 2022-07-25 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cell-surface protein YwfG of Lactococcus lactis binds to alpha-1,2-linked mannose.
Plos One, 18, 2023
|
|
7YL4
 
 | | Cell surface protein YwfG protein (apo form) | | Descriptor: | CALCIUM ION, GRAM_POS_ANCHORING domain-containing protein, SULFATE ION | | Authors: | Tsuchiya, W, Fujimoto, Z, Suzuki, C. | | Deposit date: | 2022-07-25 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cell-surface protein YwfG of Lactococcus lactis binds to alpha-1,2-linked mannose.
Plos One, 18, 2023
|
|
1VEC
 
 | | Crystal structure of the N-terminal domain of rck/p54, a human DEAD-box protein | | Descriptor: | ATP-dependent RNA helicase p54, L(+)-TARTARIC ACID, ZINC ION | | Authors: | Hogetsu, K, Matsui, T, Yukihiro, Y, Tanaka, M, Sato, T, Kumasaka, T, Tanaka, N. | | Deposit date: | 2004-03-29 | | Release date: | 2004-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural insight of human DEAD-box protein rck/p54 into its substrate recognition with conformational changes
Genes Cells, 11, 2006
|
|
3CPP
 
 | |
1OFV
 
 | | FLAVODOXIN FROM ANACYSTIS NIDULANS: REFINEMENT OF TWO FORMS OF THE OXIDIZED PROTEIN | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Smith, W.W, Pattridge, K.A, Luschinsky, C.L, Ludwig, M.L. | | Deposit date: | 1992-06-22 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined structures of oxidized flavodoxin from Anacystis nidulans.
J.Mol.Biol., 294, 1999
|
|
