2GOP
 
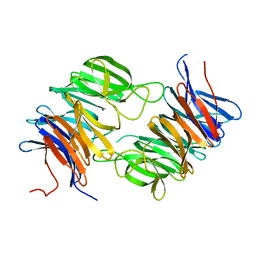 | | The beta-propeller domain of the Trilobed protease from Pyrococcus furiosus reveals an open velcro topology | | 分子名称: | Trilobed Protease | | 著者 | Bosch, J, Tamura, T, Tamura, N, Baumeister, W, Essen, L.-O. | | 登録日 | 2006-04-13 | | 公開日 | 2007-01-23 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The beta-propeller domain of the trilobed protease from Pyrococcus furiosus reveals an open Velcro topology.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2ZK7
 
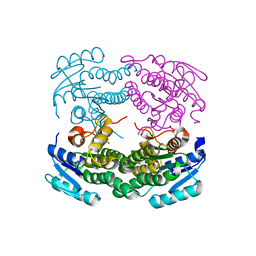 | | Structure of a C-terminal deletion mutant of Thermoplasma acidophilum aldohexose dehydrogenase (AldT) | | 分子名称: | Glucose 1-dehydrogenase related protein | | 著者 | Nishioka, T, Yasutake, Y, Nishiya, Y, Tamura, N, Tamura, T. | | 登録日 | 2008-03-12 | | 公開日 | 2009-01-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | C-terminal tail derived from the neighboring subunit is critical for the activity of Thermoplasma acidophilum D-aldohexose dehydrogenase
Proteins, 74, 2009
|
|
4ZHR
 
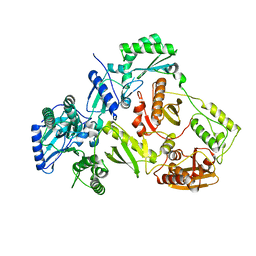 | | Structure of HIV-1 RT Q151M mutant | | 分子名称: | RT p51 subunit, RT p66 subunit | | 著者 | Nakamura, A, Tamura, N, Yasutake, Y. | | 登録日 | 2015-04-27 | | 公開日 | 2015-11-11 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.601 Å) | | 主引用文献 | Structure of the HIV-1 reverse transcriptase Q151M mutant: insights into the inhibitor resistance of HIV-1 reverse transcriptase and the structure of the nucleotide-binding pocket of Hepatitis B virus polymerase.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
2DTX
 
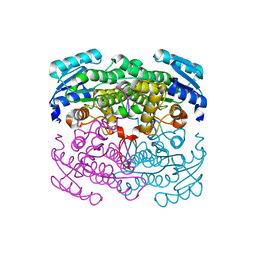 | | Structure of Thermoplasma acidophilum aldohexose dehydrogenase (AldT) in complex with D-mannose | | 分子名称: | Glucose 1-dehydrogenase related protein, SULFATE ION, beta-D-mannopyranose | | 著者 | Yasutake, Y, Nishiya, Y, Tamura, N, Tamura, T. | | 登録日 | 2006-07-18 | | 公開日 | 2007-03-27 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural Insights into Unique Substrate Selectivity of Thermoplasma acidophilumd-Aldohexose Dehydrogenase
J.Mol.Biol., 367, 2007
|
|
2DTD
 
 | | Structure of Thermoplasma acidophilum aldohexose dehydrogenase (AldT) in ligand-free form | | 分子名称: | Glucose 1-dehydrogenase related protein, SULFATE ION | | 著者 | Yasutake, Y, Nishiya, Y, Tamura, N, Tamura, T. | | 登録日 | 2006-07-12 | | 公開日 | 2007-03-27 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Insights into Unique Substrate Selectivity of Thermoplasma acidophilumd-Aldohexose Dehydrogenase
J.Mol.Biol., 367, 2007
|
|
2DTE
 
 | | Structure of Thermoplasma acidophilum aldohexose dehydrogenase (AldT) in complex with NADH | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glucose 1-dehydrogenase related protein | | 著者 | Yasutake, Y, Nishiya, Y, Tamura, N, Tamura, T. | | 登録日 | 2006-07-12 | | 公開日 | 2007-03-27 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural Insights into Unique Substrate Selectivity of Thermoplasma acidophilumd-Aldohexose Dehydrogenase
J.Mol.Biol., 367, 2007
|
|
7DBN
 
 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/M184V/F160M:DNA:dCTP ternary complex | | 分子名称: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | 著者 | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | 登録日 | 2020-10-21 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Biochemical and Structural Properties of Entecavir-Resistant Hepatitis B Virus Polymerase with L180M/M204V Mutations.
J.Virol., 95, 2021
|
|
7DBM
 
 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/M184V:DNA:dGTP ternary complex | | 分子名称: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | 著者 | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | 登録日 | 2020-10-21 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Biochemical and Structural Properties of Entecavir-Resistant Hepatitis B Virus Polymerase with L180M/M204V Mutations.
J.Virol., 95, 2021
|
|
6IKA
 
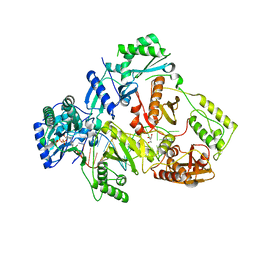 | | HIV-1 reverse transcriptase with Q151M/G112S/D113A/Y115F/F116Y/F160L/I159L:DNA:entecavir-triphosphate ternary complex | | 分子名称: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | 著者 | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | 登録日 | 2018-10-15 | | 公開日 | 2019-01-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.598 Å) | | 主引用文献 | Active-site deformation in the structure of HIV-1 RT with HBV-associated septuple amino acid substitutions rationalizes the differential susceptibility of HIV-1 and HBV against 4'-modified nucleoside RT inhibitors.
Biochem. Biophys. Res. Commun., 509, 2019
|
|
6IK9
 
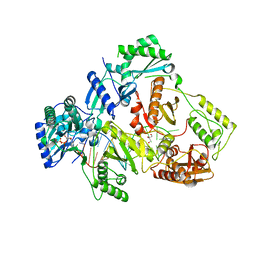 | | HIV-1 reverse transcriptase with Q151M/G112S/D113A/Y115F/F116Y/F160L/I159L:DNA:dGTP ternary complex | | 分子名称: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | 著者 | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | 登録日 | 2018-10-15 | | 公開日 | 2019-01-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.435 Å) | | 主引用文献 | Active-site deformation in the structure of HIV-1 RT with HBV-associated septuple amino acid substitutions rationalizes the differential susceptibility of HIV-1 and HBV against 4'-modified nucleoside RT inhibitors.
Biochem. Biophys. Res. Commun., 509, 2019
|
|
6KDM
 
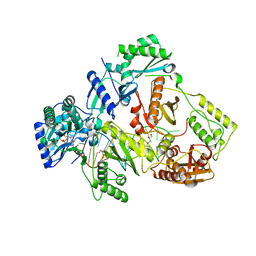 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y:DNA:entecavir 5'-triphosphate ternary complex | | 分子名称: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | 著者 | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | 登録日 | 2019-07-02 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
6KDN
 
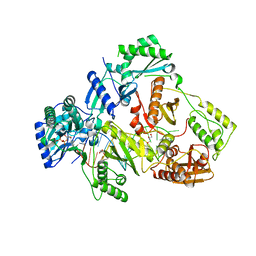 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y:DNA:dGTP ternary complex | | 分子名称: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | 著者 | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | 登録日 | 2019-07-02 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.303 Å) | | 主引用文献 | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
6KDJ
 
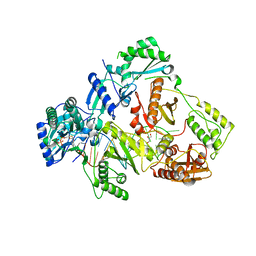 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y:DNA:lamivudine 5'-triphosphate ternary complex | | 分子名称: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | 著者 | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | 登録日 | 2019-07-02 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
6KDO
 
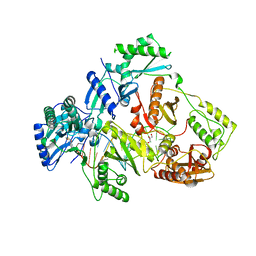 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y/M184V/F160M:DNA:lamivudine 5'-triphosphate ternary complex | | 分子名称: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | 著者 | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | 登録日 | 2019-07-02 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.573 Å) | | 主引用文献 | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
6KDK
 
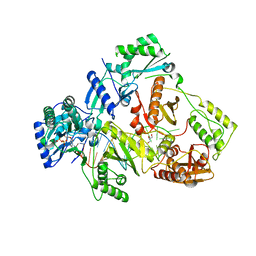 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y:DNA:dCTP ternary complex | | 分子名称: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | 著者 | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | 登録日 | 2019-07-02 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
5XN2
 
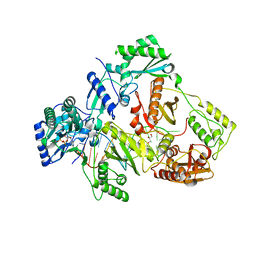 | | HIV-1 reverse transcriptase Q151M:DNA:dGTP ternary complex | | 分子名称: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 38-MER DNA aptamer, GLYCEROL, ... | | 著者 | Yasutake, Y, Tamura, N, Hayashi, H, Maeda, K. | | 登録日 | 2017-05-17 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.381 Å) | | 主引用文献 | HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir.
Sci Rep, 8, 2018
|
|
5XN0
 
 | | HIV-1 reverse transcriptase Q151M:DNA binary complex | | 分子名称: | 38-MER DNA aptamer, GLYCEROL, Pol protein, ... | | 著者 | Yasutake, Y, Tamura, N, Hayashi, H, Maeda, K. | | 登録日 | 2017-05-17 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.596 Å) | | 主引用文献 | HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir.
Sci Rep, 8, 2018
|
|
5XN1
 
 | | HIV-1 reverse transcriptase Q151M:DNA:entecavir-triphosphate ternary complex | | 分子名称: | 38-MER DNA aptamer, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Yasutake, Y, Tamura, N, Hayashi, H, Maeda, K. | | 登録日 | 2017-05-17 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.446 Å) | | 主引用文献 | HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir.
Sci Rep, 8, 2018
|
|
3WTD
 
 | | Structure of PAXX | | 分子名称: | Uncharacterized protein C9orf142 | | 著者 | Ochi, T, Blundell, T.L. | | 登録日 | 2014-04-09 | | 公開日 | 2015-01-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair.
Science, 347, 2015
|
|
3WTF
 
 | | Structure of PAXX | | 分子名称: | Uncharacterized protein C9orf142 | | 著者 | Ochi, T, Blundell, T.L. | | 登録日 | 2014-04-09 | | 公開日 | 2015-01-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.451 Å) | | 主引用文献 | DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair.
Science, 347, 2015
|
|
