7CYG
 
 | | Crystal structure of a cysteine-pair mutant (Y113C-P190C) of a bacterial bile acid transporter before disulfide bond formation | | Descriptor: | Transporter, sodium/bile acid symporter family | | Authors: | Wang, X, Lyu, Y, Ji, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-09-03 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.198 Å) | | Cite: | An engineered disulfide bridge traps and validates an outward-facing conformation in a bile acid transporter.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7CYK
 
 | | Crystal structure of a second cysteine-pair mutant (V110C-I197C) of a bacterial bile acid transporter before disulfide bond formation | | Descriptor: | MERCURY (II) ION, Transporter, sodium/bile acid symporter family | | Authors: | Wang, X, Lyu, Y, Ji, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-09-03 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.785 Å) | | Cite: | An engineered disulfide bridge traps and validates an outward-facing conformation in a bile acid transporter.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7DJC
 
 | | Crystal structure of the G26C/Q250A mutant of LeuT | | Descriptor: | LEUCINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION, ... | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-20 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
7DJ1
 
 | | Crystal structure of the G26C mutant of LeuT | | Descriptor: | LEUCINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.528 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
7DII
 
 | | Crystal structure of LeuT in lipidic cubic phase at pH 7 | | Descriptor: | LEUCINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
7DIX
 
 | | Crystal structure of LeuT in lipidic cubic phase at pH 5 | | Descriptor: | Na(+):neurotransmitter symporter (Snf family), SELENOMETHIONINE, SODIUM ION | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
7DJ2
 
 | | Crystal structure of the G26C/E290S mutant of LeuT | | Descriptor: | LEUCINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION, ... | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
6LGY
 
 | | Crystal structure of a cysteine-pair mutant (P10C-S291C) of a bacterial bile acid transporter in an inward-facing state complexed with glycine and sodium | | Descriptor: | 2,3-dihydroxypropyl (9Z)-octadec-9-enoate, GLYCINE, NONAETHYLENE GLYCOL, ... | | Authors: | Wang, X, Lyu, Y, Ji, Y, Sun, Z, Zhou, X. | | Deposit date: | 2019-12-06 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | Substrate binding in the bile acid transporter ASBT Yf from Yersinia frederiksenii.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6LH0
 
 | | Crystal structure of a cysteine-pair mutant (P10C-S291C) of a bacterial bile acid transporter in an inward-facing apo-state | | Descriptor: | 2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Transporter, sodium/bile acid symporter family | | Authors: | Wang, X, Lyu, Y, Ji, Y, Sun, Z, Zhou, X. | | Deposit date: | 2019-12-06 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.812 Å) | | Cite: | Substrate binding in the bile acid transporter ASBT Yf from Yersinia frederiksenii.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6LGZ
 
 | | Crystal structure of a cysteine-pair mutant (P10C-S291C) of a bacterial bile acid transporter in an inward-facing state complexed with sulfate | | Descriptor: | 2,3-dihydroxypropyl (9Z)-octadec-9-enoate, SULFATE ION, Transporter, ... | | Authors: | Wang, X, Lyu, Y, Ji, Y, Sun, Z, Zhou, X. | | Deposit date: | 2019-12-06 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.428 Å) | | Cite: | Substrate binding in the bile acid transporter ASBT Yf from Yersinia frederiksenii.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6LH1
 
 | | Crystal structure of a cysteine-pair mutant (Y113C-P190C) of a bacterial bile acid transporter trapped in an outward-facing conformation | | Descriptor: | 2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CITRIC ACID, Transporter, ... | | Authors: | Wang, X, Lyu, Y, Ji, Y, Sun, Z, Zhou, X. | | Deposit date: | 2019-12-06 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.861 Å) | | Cite: | An engineered disulfide bridge traps and validates an outward-facing conformation in a bile acid transporter.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6LGV
 
 | | Crystal structure of a cysteine-pair mutant (P10C-S291C) of a bacterial bile acid transporter in an inward-facing state complexed with citrate | | Descriptor: | 2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CITRIC ACID, Transporter, ... | | Authors: | Wang, X, Lyu, Y, Ji, Y, Sun, Z, Zhou, X. | | Deposit date: | 2019-12-06 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Substrate binding in the bile acid transporter ASBT Yf from Yersinia frederiksenii.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6LQX
 
 | | Crystal structure of the CBP bromodomain in complex with small molecule LC-CPin7 | | Descriptor: | (1~{S},6~{R})-6-[(1-methoxycarbonyl-3,4-dihydro-2~{H}-quinolin-6-yl)carbamoyl]cyclohex-3-ene-1-carboxylic acid, CREB-binding protein, SODIUM ION | | Authors: | Chen, Y, Zhang, F, Sun, Z, Bi, X, Luo, C. | | Deposit date: | 2020-01-14 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Design, synthesis and biological evaluation of novel small molecule inhibitor of the CBP bromodomain with possible anti-leukemia effects
To Be Published
|
|
7Y96
 
 | |
7Y9B
 
 | | Crystal structure of the membrane (M) protein of a SARS-COV-2-related coronavirus | | Descriptor: | 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, Membrane protein | | Authors: | Wang, X, Sun, Z, Zhou, X. | | Deposit date: | 2022-06-24 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.214 Å) | | Cite: | Crystal structure of the membrane (M) protein from a bat betacoronavirus.
Pnas Nexus, 2, 2023
|
|
7VFA
 
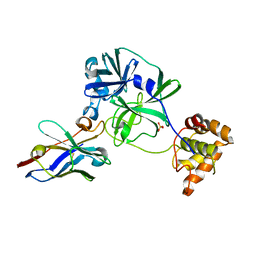 | | the complex of SARS-CoV2 3CL and NB1A2 | | Descriptor: | 3C-like proteinase, NB1A2, SULFATE ION | | Authors: | Sun, Z.C, Wang, L, Geng, Y. | | Deposit date: | 2021-09-11 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An extended conformation of SARS-CoV-2 main protease reveals allosteric targets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VFB
 
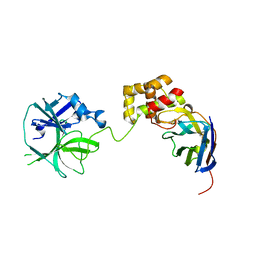 | | the complex of SARS-CoV2 3cl and NB2B4 | | Descriptor: | 3C-like proteinase, nb2b4 | | Authors: | Geng, Y, Sun, Z.C, Wang, L. | | Deposit date: | 2021-09-11 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An extended conformation of SARS-CoV-2 main protease reveals allosteric targets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5WUU
 
 | |
5YAO
 
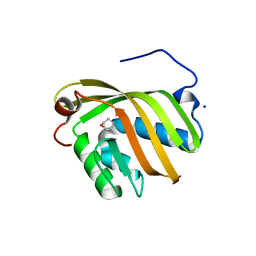 | | The complex structure of SZ529 and expoxid | | Descriptor: | (1R,5S)-6-oxabicyclo[3.1.0]hexane, Limonene-1,2-epoxide hydrolase, SODIUM ION | | Authors: | Lian, W, Sun, Z.T, Zhou, J.H, Reetz, M.T. | | Deposit date: | 2017-09-01 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.611 Å) | | Cite: | Structural and Computational Insight into the Catalytic Mechanism of Limonene Epoxide Hydrolase Mutants in Stereoselective Transformations
J. Am. Chem. Soc., 140, 2018
|
|
5YNG
 
 | | Crystal structure of SZ348 in complex with cyclopentene oxide | | Descriptor: | (1R,5S)-6-oxabicyclo[3.1.0]hexane, Limonene-1,2-epoxide hydrolase, NICKEL (II) ION, ... | | Authors: | Wu, L, Sun, Z.T, Reetz, M.T, Zhou, J.H. | | Deposit date: | 2017-10-24 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Structural and Computational Insight into the Catalytic Mechanism of Limonene Epoxide Hydrolase Mutants in Stereoselective Transformations.
J. Am. Chem. Soc., 140, 2018
|
|
5YQT
 
 | | Crystal Structure of the L74F/M78V/I80V/L114F mutant of LEH complexed with cyclopentene oxide | | Descriptor: | (1R,5S)-6-oxabicyclo[3.1.0]hexane, Limonene-1,2-epoxide hydrolase | | Authors: | Kong, X.D, Sun, Z.T, Wu, L, Reetz, M.T, Zhou, J.H, Xu, J.H. | | Deposit date: | 2017-11-07 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Computational Insight into the Catalytic Mechanism of Limonene Epoxide Hydrolase Mutants in Stereoselective Transformations.
J. Am. Chem. Soc., 140, 2018
|
|
4IKG
 
 | |
6KX2
 
 | | Crystal structure of GDP bound RhoA protein | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Transforming protein RhoA | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2019-09-09 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.454 Å) | | Cite: | Covalent Inhibitors Allosterically Block the Activation of Rho Family Proteins and Suppress Cancer Cell Invasion.
Adv Sci, 7, 2020
|
|
6KX3
 
 | | Crystal structure of RhoA protein with covalent inhibitor DC-Rhoin | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Transforming protein RhoA, prop-2-enyl (3R)-1,1-bis(oxidanylidene)-2,3-dihydro-1-benzothiophene-3-carboxylate | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2019-09-09 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Covalent Inhibitors Allosterically Block the Activation of Rho Family Proteins and Suppress Cancer Cell Invasion.
Adv Sci, 7, 2020
|
|
5QTT
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(3R,7S)-7-{[5-amino-1-(3-chloro-2-fluorophenyl)-1H-pyrazole-4-carbonyl]amino}-3-methyl-2-oxo-2,3,4,5,6,7-hexahydro-1H-12,8-(metheno)-1,9-benzodiazacyclotetradecin-15-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2019-10-16 | | Release date: | 2019-12-25 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Potent, Orally Bioavailable and Efficacious Macrocyclic Inhibitors of Factor XIa. Discovery of Pyridine-Based Macrocycles Possessing Phenylazole Carboxamide P1 Groups.
J.Med.Chem., 63, 2019
|
|
