6ULN
 
 | |
6UON
 
 | |
1FDO
 
 | | OXIDIZED FORM OF FORMATE DEHYDROGENASE H FROM E. COLI | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FORMATE DEHYDROGENASE H, IRON/SULFUR CLUSTER, ... | | Authors: | Sun, P.D, Boyington, J.C. | | Deposit date: | 1997-01-27 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster.
Science, 275, 1997
|
|
1FDI
 
 | | OXIDIZED FORM OF FORMATE DEHYDROGENASE H FROM E. COLI COMPLEXED WITH THE INHIBITOR NITRITE | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FORMATE DEHYDROGENASE H, IRON/SULFUR CLUSTER, ... | | Authors: | Sun, P.D, Boyington, J.C. | | Deposit date: | 1997-01-28 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster.
Science, 275, 1997
|
|
1AA6
 
 | | REDUCED FORM OF FORMATE DEHYDROGENASE H FROM E. COLI | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FORMATE DEHYDROGENASE H, IRON/SULFUR CLUSTER, ... | | Authors: | Sun, P.D, Boyington, J.C. | | Deposit date: | 1997-01-23 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster.
Science, 275, 1997
|
|
6ULR
 
 | |
6ULI
 
 | |
6ULK
 
 | |
2DLI
 
 | | KILLER IMMUNOGLOBULIN RECEPTOR 2DL2,TRIGONAL FORM | | Descriptor: | PROTEIN (MHC CLASS I NK CELL RECEPTOR PRECURSOR) | | Authors: | Sun, P.D, Snyder, G.A. | | Deposit date: | 1999-03-08 | | Release date: | 2000-01-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the HLA-Cw3 allotype-specific killer cell inhibitory receptor KIR2DL2
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3KHQ
 
 | | Crystal structure of murine Ig-beta (CD79b) in the monomeric form | | Descriptor: | B-cell antigen receptor complex-associated protein beta chain, CITRATE ANION, GLUTATHIONE, ... | | Authors: | Radaev, S, Sun, P.D. | | Deposit date: | 2009-10-30 | | Release date: | 2010-08-25 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Studies of Igalphabeta and Its Assembly with the B Cell Antigen Receptor.
Structure, 18, 2010
|
|
1EFX
 
 | | STRUCTURE OF A COMPLEX BETWEEN THE HUMAN NATURAL KILLER CELL RECEPTOR KIR2DL2 AND A CLASS I MHC LIGAND HLA-CW3 | | Descriptor: | BETA-2-MICROGLOBULIN, HLA-CW3 (HEAVY CHAIN), NATURAL KILLER CELL RECEPTOR KIR2DL2, ... | | Authors: | Boyington, J.C, Motyka, S.A, Schuck, P, Brooks, A.G, Sun, P.D. | | Deposit date: | 2000-02-10 | | Release date: | 2000-06-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand.
Nature, 405, 2000
|
|
4X4M
 
 | | Structure of FcgammaRI in complex with Fc reveals the importance of glycan recognition for high affinity IgG binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lu, J, Sun, P.D. | | Deposit date: | 2014-12-03 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.485 Å) | | Cite: | Structure of Fc gamma RI in complex with Fc reveals the importance of glycan recognition for high-affinity IgG binding.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3RJD
 
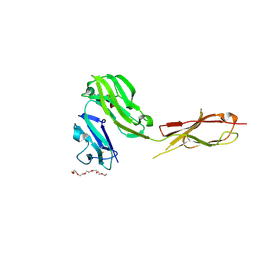 | | Crystal structure of Fc RI and its implication to high affinity immunoglobulin G binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ... | | Authors: | Lu, J, Sun, P.D. | | Deposit date: | 2011-04-15 | | Release date: | 2011-09-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of Fc gamma receptor I and its implication in high affinity gamma-immunoglobulin binding.
J.Biol.Chem., 286, 2011
|
|
1FNL
 
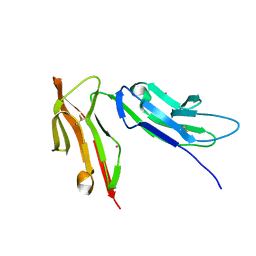 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR DOMAIN OF A HUMAN FCGRIII | | Descriptor: | LOW AFFINITY IMMUNOGLOBULIN GAMMA FC REGION RECEPTOR III-B, MERCURY (II) ION | | Authors: | Zhang, Y, Boesen, C.C, Radaev, S, Brooks, A.G, Fridman, W.H, Sautes-Fridman, C, Sun, P.D. | | Deposit date: | 2000-08-22 | | Release date: | 2000-11-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the extracellular domain of a human Fc gamma RIII.
Immunity, 13, 2000
|
|
2ZG3
 
 | |
7SU9
 
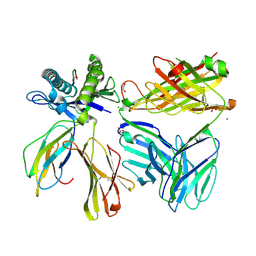 | |
2ZG1
 
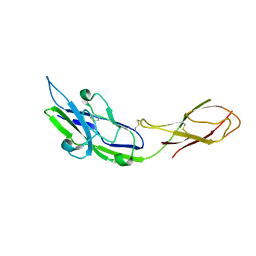 | |
2ZG2
 
 | |
8VJZ
 
 | | HLA-A*03:01 with WT KRAS-10mer | | Descriptor: | Beta-2-microglobulin, GLYCEROL, GTPase KRas, ... | | Authors: | Sim, M.J.W, Sun, P.D. | | Deposit date: | 2024-01-08 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and structural characterization of a mutant KRAS-G12V specific TCR restricted by HLA-A3.
Eur.J.Immunol., 54, 2024
|
|
1B6E
 
 | | HUMAN CD94 | | Descriptor: | CD94 | | Authors: | Boyington, J.C, Riaz, A.N, Patamawenu, A, Coligan, J.E, Brooks, A.G, Sun, P.D. | | Deposit date: | 1999-01-14 | | Release date: | 1999-06-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of CD94 reveals a novel C-type lectin fold: implications for the NK cell-associated CD94/NKG2 receptors.
Immunity, 10, 1999
|
|
3D5O
 
 | | Structural recognition and functional activation of FcrR by innate pentraxins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Low affinity immunoglobulin gamma Fc region receptor II-a, ... | | Authors: | Lu, J, Marnell, L.L, Marjon, K.D, Mold, C, Du Clos, T.W, Sun, P.D. | | Deposit date: | 2008-05-16 | | Release date: | 2008-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural recognition and functional activation of FcgammaR by innate pentraxins.
Nature, 456, 2008
|
|
3KG5
 
 | | Crystal structure of human Ig-beta homodimer | | Descriptor: | B-cell antigen receptor complex-associated protein beta chain | | Authors: | Radaev, S, Sun, P.D. | | Deposit date: | 2009-10-28 | | Release date: | 2010-08-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and Functional Studies of Igalphabeta and Its Assembly with the B Cell Antigen Receptor.
Structure, 18, 2010
|
|
3KHO
 
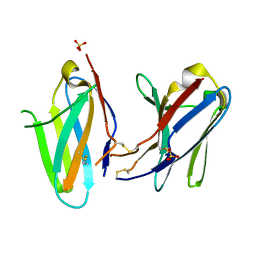 | | Crystal structure of murine Ig-beta (CD79b) homodimer | | Descriptor: | B-cell antigen receptor complex-associated protein beta chain, SULFATE ION | | Authors: | Radaev, S, Sun, P.D. | | Deposit date: | 2009-10-30 | | Release date: | 2010-08-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural and Functional Studies of Igalphabeta and Its Assembly with the B Cell Antigen Receptor.
Structure, 18, 2010
|
|
8RRO
 
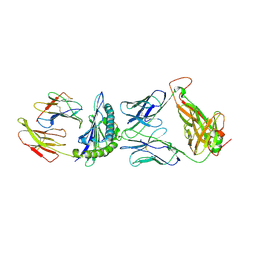 | | G12V-TCR complex with HLA-A3 | | Descriptor: | Beta-2-microglobulin, G12V-TCR alpha chain, G12V-TCR beta chain, ... | | Authors: | Sim, M.J.W, Sun, P.D. | | Deposit date: | 2024-01-23 | | Release date: | 2024-11-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Identification and structural characterization of a mutant KRAS-G12V specific TCR restricted by HLA-A3.
Eur.J.Immunol., 54, 2024
|
|
6C68
 
 | | MHC-independent t cell receptor A11 | | Descriptor: | T-cell receptor alpha chain, T-cell receptor beta chain | | Authors: | Lu, J, Van Laethem, F, Saba, I, Chu, J, Bhattacharya, A, Love, N.C, Tikhonova, A, Radaev, S, Sun, X, Ko, A, Arnon, T, Shifrut, E, Friedman, N, Weng, N, Singer, A, Sun, P.D. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of MHC-Independent TCRs and Their Recognition of Native Antigen CD155.
J Immunol., 204, 2020
|
|
