6MGJ
 
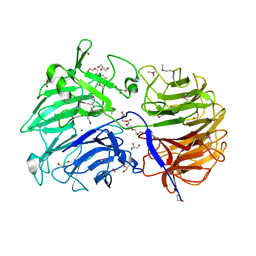 | | Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, apoenzyme | | 分子名称: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, GLYCEROL, ... | | 著者 | Stogios, P.J, Skarina, T, Nocek, B, Arnal, G, Brumer, H, Savchenko, A. | | 登録日 | 2018-09-14 | | 公開日 | 2019-01-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural enzymology reveals the molecular basis of substrate regiospecificity and processivity of an exemplar bacterial glycoside hydrolase family 74endo-xyloglucanase.
Biochem. J., 475, 2018
|
|
6MSW
 
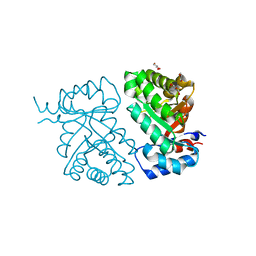 | | Crystal structure of BH1352 2-deoxyribose-5-phosphate from Bacillus halodurans, K184L mutant | | 分子名称: | Deoxyribose-phosphate aldolase, GLYCEROL | | 著者 | Stogios, P.J, Skarina, T, Kim, T, Yim, V, Yakunin, A, Savchenko, A. | | 登録日 | 2018-10-18 | | 公開日 | 2019-10-23 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.169 Å) | | 主引用文献 | Rational engineering of 2-deoxyribose-5-phosphate aldolases for the biosynthesis of (R)-1,3-butanediol.
J.Biol.Chem., 295, 2020
|
|
6MGK
 
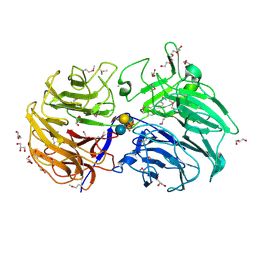 | | Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, in complex with XLX xyloglucan | | 分子名称: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, GLYCEROL, ... | | 著者 | Stogios, P.J, Skarina, T, Nocek, B, Arnal, G, Brumer, H, Savchenko, A. | | 登録日 | 2018-09-14 | | 公開日 | 2019-01-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural enzymology reveals the molecular basis of substrate regiospecificity and processivity of an exemplar bacterial glycoside hydrolase family 74endo-xyloglucanase.
Biochem. J., 475, 2018
|
|
6NPS
 
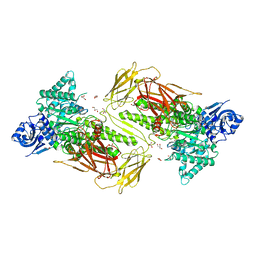 | | Crystal structure of GH115 enzyme AxyAgu115A from Amphibacillus xylanus | | 分子名称: | AxyAgu115A, CHLORIDE ION, GLYCEROL | | 著者 | Stogios, P.J, Skarina, T, Di Leo, R, Yan, R, Master, E, Savchenko, A. | | 登録日 | 2019-01-18 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structural characterization of the family GH115 alpha-glucuronidase from Amphibacillus xylanus yields insight into its coordinated action with alpha-arabinofuranosidases.
N Biotechnol, 2021
|
|
4WZ2
 
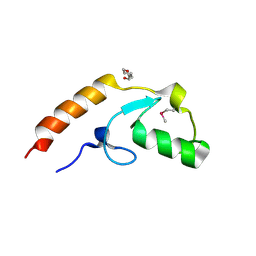 | | Crystal structure of U-box 2 of LubX / LegU2 / Lpp2887 from Legionella pneumophila str. Paris, Ile175Met mutant | | 分子名称: | CHLORIDE ION, E3 ubiquitin-protein ligase LubX, HEXANE-1,6-DIOL | | 著者 | Stogios, P.J, Qualie, A.T, Skarina, T, Nocek, B, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-11-18 | | 公開日 | 2015-01-28 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (3.408 Å) | | 主引用文献 | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
4WZ3
 
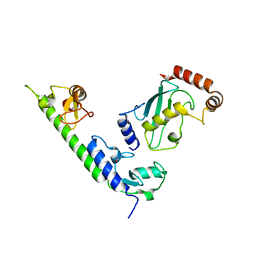 | | Crystal structure of the complex between LubX/LegU2/Lpp2887 U-box 1 and Homo sapiens UBE2D2 | | 分子名称: | E3 ubiquitin-protein ligase LubX, Ubiquitin-conjugating enzyme E2 D2 | | 著者 | Stogios, P.J, Quaile, A.T, Skarina, T, Nocek, B, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-11-18 | | 公開日 | 2015-01-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
4WZ0
 
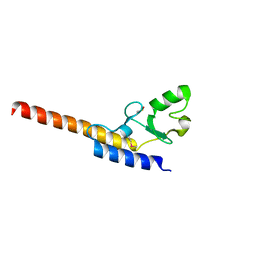 | | Crystal structure of U-box 1 of LubX / LegU2 / Lpp2887 from Legionella pneumophila str. Paris | | 分子名称: | E3 ubiquitin-protein ligase LubX | | 著者 | Stogios, P.J, Quaile, A.T, Skarina, T, Stein, A, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-11-18 | | 公開日 | 2015-01-14 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (1.954 Å) | | 主引用文献 | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
7TQ1
 
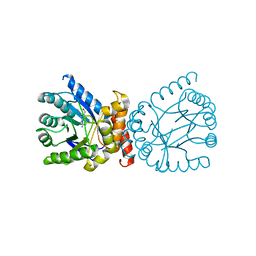 | | Crystal structure of adaptive laboratory evolved sulfonamide-resistant Dihydropteroate Synthase (DHPS) from Escherichia coli in complex with 6-hydroxymethylpterin | | 分子名称: | 6-HYDROXYMETHYLPTERIN, Dihydropteroate synthase | | 著者 | Stogios, P.J, Skarina, T, Tan, K, Venkatesan, M, Fruci, M, Joachimiak, A, Savchenko, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-01-26 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.73 Å) | | 主引用文献 | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
4XA9
 
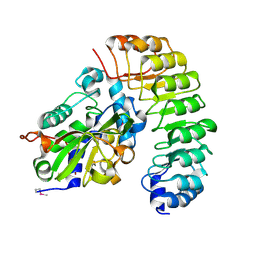 | | Crystal structure of the complex between the N-terminal domain of RavJ and LegL1 from Legionella pneumophila str. Philadelphia | | 分子名称: | Gala protein type 1, 3 or 4, Uncharacterized protein | | 著者 | Stogios, P.J, Cuff, M.E, Nocek, B, Evdokimova, E, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-12-13 | | 公開日 | 2015-01-28 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila.
Mol. Syst. Biol., 12, 2016
|
|
4XI1
 
 | | Crystal structure of U-box 2 of LubX / LegU2 / Lpp2887 from Legionella pneumophila str. Paris, wild-type | | 分子名称: | CHLORIDE ION, E3 ubiquitin-protein ligase LubX, GLYCEROL, ... | | 著者 | Stogios, P.J, Quaile, T, Skarina, T, Cuff, M, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-01-06 | | 公開日 | 2015-01-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.983 Å) | | 主引用文献 | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
7UUN
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with neomycin | | 分子名称: | 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, NEOMYCIN | | 著者 | Stogios, P.J, Evdokimova, E, Di Leo, R, Osipiuk, J, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2022-04-28 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
7UUM
 
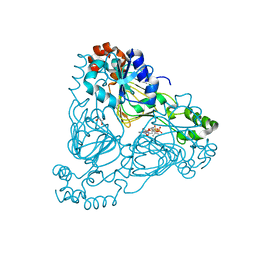 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with paromomycin and coenzyme A | | 分子名称: | Aminocyclitol acetyltransferase ApmA, COENZYME A, GLYCEROL, ... | | 著者 | Stogios, P.J, Evdokimova, E, Osipiuk, J, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2022-04-28 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.74 Å) | | 主引用文献 | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
7UUL
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with kanamycin B and coenzyme A | | 分子名称: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, ... | | 著者 | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2022-04-28 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
7UUO
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA H135A mutant, complex with tobramycin and coenzyme A | | 分子名称: | 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, COENZYME A, ... | | 著者 | Stogios, P.J, Evdokimova, E, Michalska, K, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2022-04-28 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
7UUK
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with tobramycin | | 分子名称: | Aminocyclitol acetyltransferase ApmA, CHLORIDE ION, TOBRAMYCIN | | 著者 | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2022-04-28 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.82 Å) | | 主引用文献 | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
4MPH
 
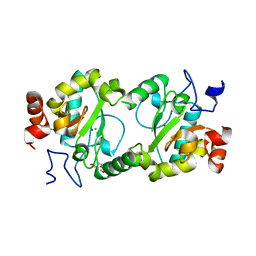 | | Crystal structure of BaLdcB / VanY-like L,D-carboxypeptidase Zinc(II)-bound | | 分子名称: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, D-alanyl-D-alanine carboxypeptidase family protein, ... | | 著者 | Stogios, P.J, Wawrzak, Z, Onopriyenko, O, Skarina, T, Shatsman, S, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-09-12 | | 公開日 | 2013-09-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.0301 Å) | | 主引用文献 | Structure of the LdcB LD-Carboxypeptidase Reveals the Molecular Basis of Peptidoglycan Recognition.
Structure, 22, 2014
|
|
4MUT
 
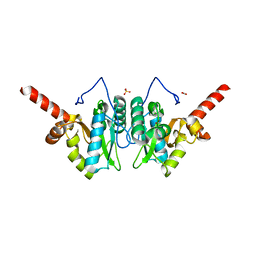 | | Crystal structure of vancomycin resistance D,D-dipeptidase/D,D-pentapeptidase VanXYc D59S mutant in complex with D-Alanine | | 分子名称: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, D,D-dipeptidase/D,D-carboxypeptidase, ... | | 著者 | Stogios, P.J, Evdokimova, E, Meziane-Cherif, D, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-09-23 | | 公開日 | 2013-10-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MUS
 
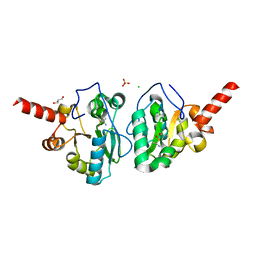 | | Crystal structure of vancomycin resistance D,D-dipeptidase/D,D-pentapeptidase VanXYc D59S mutant in complex with D-Ala-D-Ala phosphinate analog | | 分子名称: | (2R)-3-[(R)-[(1R)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, (2R)-3-[(R)-[(1S)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, CHLORIDE ION, ... | | 著者 | Stogios, P.J, Evdokimova, E, Meziane-Cherif, D, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-09-23 | | 公開日 | 2013-10-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.675 Å) | | 主引用文献 | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MUR
 
 | | Crystal structure of vancomycin resistance D,D-dipeptidase/D,D-pentapeptidase VanXYc D59S mutant | | 分子名称: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, D,D-dipeptidase/D,D-carboxypeptidase, ... | | 著者 | Stogios, P.J, Evdokimova, E, Meziane-Cherif, D, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-09-23 | | 公開日 | 2013-10-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MUQ
 
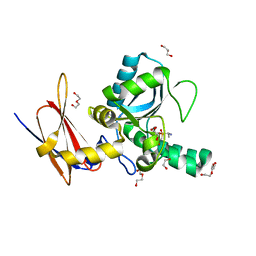 | | Crystal Structure of Vancomycin Resistance D,D-dipeptidase VanXYg in complex with D-Ala-D-Ala phosphinate analog | | 分子名称: | (2R)-3-[(R)-[(1R)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, (2R)-3-[(R)-[(1S)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, 1,2-ETHANEDIOL, ... | | 著者 | Stogios, P.J, Evdokimova, E, Meziane-Cherif, D, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-09-23 | | 公開日 | 2013-10-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.364 Å) | | 主引用文献 | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6CD7
 
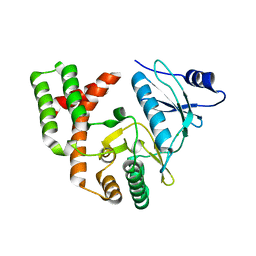 | | Crystal structure of APH(2")-IVa in complex with plazomicin | | 分子名称: | (2S)-4-amino-N-[(1R,2S,3S,4R,5S)-5-amino-4-{[(2S,3R)-3-amino-6-{[(2-hydroxyethyl)amino]methyl}-3,4-dihydro-2H-pyran-2-y l]oxy}-2-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranosyl]oxy}-3-hydroxycyclohexyl]-2-hydroxybutanamide, APH(2'')-Id, CHLORIDE ION | | 著者 | Stogios, P.J, Evdokimova, E, Dong, A, Di Leo, R, Savchenko, A, Satchell, K.J, Joachimiak, J, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-02-08 | | 公開日 | 2018-02-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Plazomicin Retains Antibiotic Activity against Most Aminoglycoside Modifying Enzymes.
ACS Infect Dis, 4, 2018
|
|
6D33
 
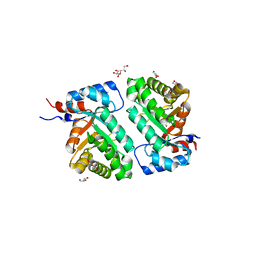 | | Crystal structure of BH1352 2-deoxyribose-5-phosphate from Bacillus halodurans | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyribose-phosphate aldolase, GLYCEROL | | 著者 | Stogios, P.J, Skarina, T, Kim, T, Yim, V, Yakunin, A, Savchenko, A. | | 登録日 | 2018-04-14 | | 公開日 | 2019-10-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.502 Å) | | 主引用文献 | Rational engineering of 2-deoxyribose-5-phosphate aldolases for the biosynthesis of (R)-1,3-butanediol.
J.Biol.Chem., 295, 2020
|
|
7ROA
 
 | | Crystal structure of EntV136 from Enterococcus faecalis | | 分子名称: | EntV | | 著者 | Stogios, P.J, Evdokimova, E, Kim, Y, Garsin, D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2021-07-30 | | 公開日 | 2022-10-12 | | 最終更新日 | 2023-01-25 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Structural and functional analysis of EntV reveals a 12 amino acid fragment protective against fungal infections.
Nat Commun, 13, 2022
|
|
7S2K
 
 | | Crystal structure of sulfonamide resistance enzyme Sul2 in complex with 7,8-dihydropteroate, magnesium, and pyrophosphate | | 分子名称: | 4-AMINOBENZOIC ACID, 7,8-DIHYDROPTEROATE, CHLORIDE ION, ... | | 著者 | Stogios, P.J, Skarina, T, Michalska, K, Venkatesan, M, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-09-03 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
7S2J
 
 | | Crystal structure of sulfonamide resistance enzyme Sul2 apoenzyme | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Stogios, P.J, Skarina, T, Michalska, K, Venkatesan, M, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-09-03 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
