9OZ0
 
 | | Crystal structure of T6SS effector-immunity complex PA3907-PA3908 from Pseudomonas aeruginosa | | Descriptor: | CITRIC ACID, Immunity protein 52 domain-containing protein, MAGNESIUM ION, ... | | Authors: | Stogios, P.J, Borek, D, Skarina, T, Osipiuk, J, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2025-06-04 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.871 Å) | | Cite: | Crystal structure of T6SS effector-immunity complex PA3907-PA3908 from Pseudomonas aeruginosa
To Be Published
|
|
5KL9
 
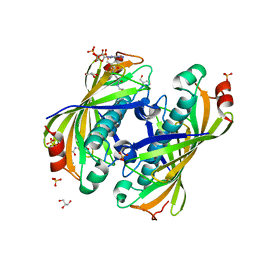 | | Crystal structure of a putative acyl-CoA thioesterase EC709/ECK0725 from Escherichia coli in complex with CoA | | Descriptor: | Acyl-CoA thioester hydrolase YbgC, COENZYME A, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-06-23 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of a putative acyl-CoA thioesterase EC709/ECK0725 from Escherichia coli in complex with CoA
To Be Published
|
|
9MTO
 
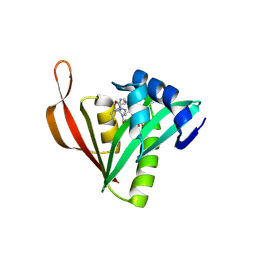 | | Crystal structure of acetyltransferase CBU0801 from Coxiella burnetii | | Descriptor: | COENZYME A, [Ribosomal protein bS18]-alanine N-acetyltransferase | | Authors: | Stogios, P.J, Evdokimova, E, Chang, C, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2025-01-11 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of acetyltransferase CBU0801 from Coxiella burnetii
To Be Published
|
|
4GKH
 
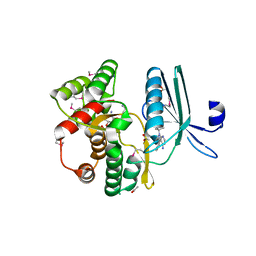 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor 1-NA-PP1 | | Descriptor: | 1-tert-butyl-3-(naphthalen-1-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Todorovic, N, Capretta, A, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-11 | | Release date: | 2012-09-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.863 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
4GKI
 
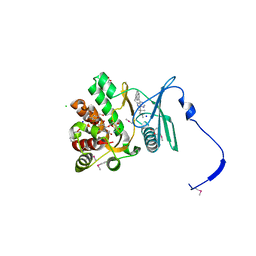 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor 1-NM-PP1 | | Descriptor: | 1-tert-butyl-3-(naphthalen-1-ylmethyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Todorovic, N, Capretta, A, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-11 | | Release date: | 2012-09-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
3V75
 
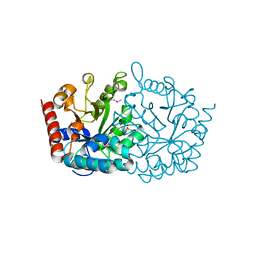 | | Crystal structure of putative orotidine 5'-phosphate decarboxylase from Streptomyces avermitilis ma-4680 | | Descriptor: | Orotidine 5'-phosphate decarboxylase | | Authors: | Stogios, P.J, Xu, X, Cui, H, Kudritska, M, Tan, K, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-12-20 | | Release date: | 2012-05-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of putative orotidine 5'-phosphate decarboxylase from Streptomyces avermitilis ma-4680
To be Published
|
|
3VCX
 
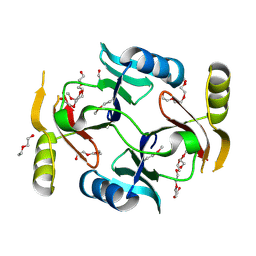 | | Crystal structure of a putative glyoxalase/bleomycin resistance protein from Rhodopseudomonas palustris CGA009 | | Descriptor: | Glyoxalase/Bleomycin resistance protein/dioxygenase domain, TETRAETHYLENE GLYCOL | | Authors: | Stogios, P.J, Chang, C, Evdokimova, E, Egorova, O, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-01-04 | | Release date: | 2012-01-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystal structure of a putative glyoxalase/bleomycin resistance protein from Rhodopseudomonas palustris CGA009
To be Published
|
|
5DT9
 
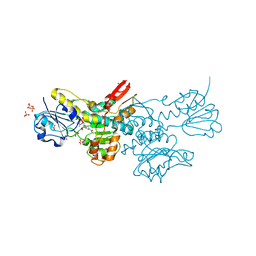 | | Crystal structure of a putative D-Erythronate-4-Phosphate Dehydrogenase from Vibrio cholerae | | Descriptor: | CHLORIDE ION, Erythronate-4-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Stogios, P.J, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-17 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.663 Å) | | Cite: | Crystal structure of a putative D-Erythronate-4-Phosphate Dehydrogenase from Vibrio cholerae
To Be Published
|
|
4WH5
 
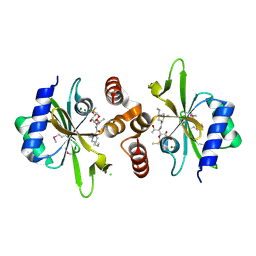 | | Crystal structure of lincosamide antibiotic adenylyltransferase LnuA, lincomycin-bound | | Descriptor: | CHLORIDE ION, LINCOMYCIN, Lincosamide resistance protein, ... | | Authors: | Stogios, P.J, Dong, A, Minasov, G, Evdokimova, E, Egorova, O, Kudritska, M, Yim, O, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-20 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | CRYSTAL STRUCTURE OF LINCOSAMIDE ANTIBIOTIC ADENYLYLTRANSFERASE LNUA, LINCOMYCIN BOUND
To Be Published
|
|
6WQB
 
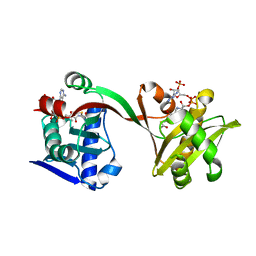 | | Crystal structure of VipF from Legionella hackeliae in complex with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, N-terminal acetyltransferase, GNAT family | | Authors: | Stogios, P.J, Skarina, T, Wawrzak, Z, Sandoval, J, Di Leo, R, Savchenko, A. | | Deposit date: | 2020-04-28 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of VipF from Legionella hackeliae in complex with acetyl-CoA
To Be Published
|
|
6WQC
 
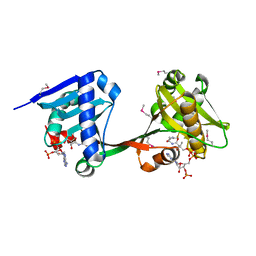 | | Crystal structure of VipF from Legionella hackeliae in complex with CoA | | Descriptor: | COENZYME A, N-terminal acetyltransferase, GNAT family | | Authors: | Stogios, P.J, Skarina, T, Wawrzak, Z, Sandoval, J, Di Leo, R, Savchenko, A. | | Deposit date: | 2020-04-28 | | Release date: | 2021-11-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of VipF from Legionella hackeliae in complex with CoA
To Be Published
|
|
8D5H
 
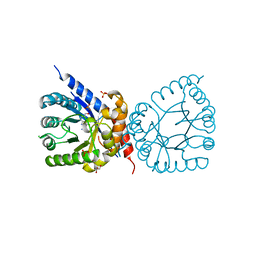 | | Crystal structure of dihydropteroate synthase (folP-SMZ_B27) from soil uncultured bacterium in complex with 6-hydroxymethyl-7,8-dihydropterin | | Descriptor: | 6-HYDROXYMETHYLPTERIN, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stogios, P.J, Skarina, T, Osipiuk, J, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-06-04 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of dihydropteroate synthase (folP-SMZ_B27) from soil uncultured bacterium in complex with 6-hydroxymethyl-7,8-dihydropterin
To Be Published
|
|
8D5I
 
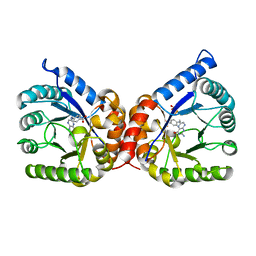 | | Crystal structure of dihydropteroate synthase H182G mutant (folP-SMZ_B27) from soil uncultured bacterium in complex with pteroic acid and pyrophosphate | | Descriptor: | PTEROIC ACID, PYROPHOSPHATE 2-, folP-SMZ_B27 | | Authors: | Stogios, P.J, Skarina, T, Kim, Y, Di Leo, R, Venkatesan, M, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-06-04 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of dihydropteroate synthase H182G mutant (folP-SMZ_B27) from soil uncultured bacterium in complex with pteroic acid
To Be Published
|
|
8D5G
 
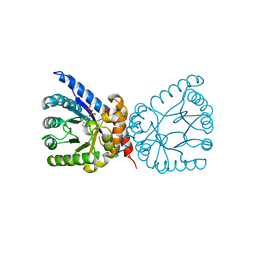 | | Crystal structure of dihydropteroate synthase (folP-SMZ_B27) from soil uncultured bacterium in complex with 6-hydroxymethyl-7,8-dihydropterin pyrophosphate | | Descriptor: | 6-HYDROXYMETHYLPTERIN-DIPHOSPHATE, CHLORIDE ION, folP-SMZ_B27 | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-06-04 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of dihydropteroate synthase (folP-SMZ_B27) from soil uncultured bacterium in complex with 6-hydroxymethyl-7,8-dihydropterin pyrophosphate
To Be Published
|
|
6M8U
 
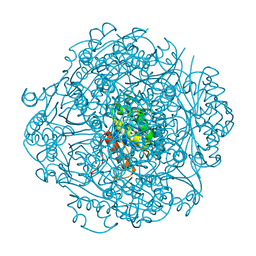 | | Crystal structure of UbiX-like FMN prenyltransferase AF1214 from Archaeoglobus fulgidus, prenylated-FMN complex | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Flavin prenyltransferase UbiX, PHOSPHATE ION | | Authors: | Stogios, P.J, Skarina, T, Khusnutidinova, A, Wawrzak, Z, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2018-08-22 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.221 Å) | | Cite: | Crystal structure of UbiX-like FMN prenyltransferase AF1214 from Archaeoglobus fulgidus, prenylated-FMN complex
To Be Published
|
|
6M8V
 
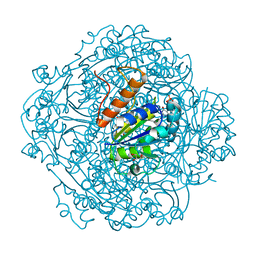 | | Crystal structure of UbiX-like FMN prenyltransferase MJ0101 from Methanocaldococcus jannaschii, FMN complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavin prenyltransferase UbiX, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Khusnutdinova, A, Wawrzak, Z, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2018-08-22 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.221 Å) | | Cite: | Crystal structure of UbiX-like FMN prenyltransferase MJ0101 from Methanocaldococcus jannaschii, FMN complex
To Be Published
|
|
6M8T
 
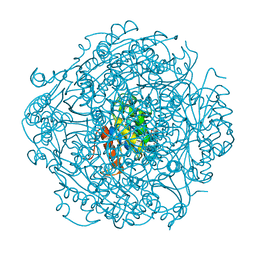 | | Crystal structure of UbiX-like FMN prenyltransferase AF1214 from Archaeoglobus fulgidus, FMN complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavin prenyltransferase UbiX, PHOSPHATE ION | | Authors: | Stogios, P.J, Skarina, T, Khusnutidnova, A, Wawrzak, Z, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2018-08-22 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of UbiX-like FMN prenyltransferase AF1214 from Archaeoglobus fulgidus, FMN complex
To Be Published
|
|
9NZL
 
 | | Crystal structure of Yck2 with DJS-02-24-06 | | Descriptor: | (4M)-4-[3-(5-methylpyridin-2-yl)-1H-pyrazol-4-yl]quinoline, CHLORIDE ION, SULFATE ION, ... | | Authors: | Stogios, P.J, Shirley, D, Cowen, L.E, Willson, T, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2025-04-01 | | Release date: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of Yck2 with DJS-02-24-06
To Be Published
|
|
9NZK
 
 | | Crystal structure of Yck2 with LY364947 | | Descriptor: | 4-(3-PYRIDIN-2-YL-1H-PYRAZOL-4-YL)QUINOLINE, CHLORIDE ION, non-specific serine/threonine protein kinase | | Authors: | Stogios, P.J, Shirley, D, Cowen, L.E, Willson, T, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2025-04-01 | | Release date: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Yck2 with LY364947
To Be Published
|
|
9P0P
 
 | | Crystal structure of 23S rRNA methyltransferase Cfr, apoenzyme | | Descriptor: | Ribosomal RNA large subunit methyltransferase Cfr | | Authors: | Stogios, P.J, Skarina, T, Kim, Y, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2025-06-07 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of 23S rRNA methyltransferase Cfr, apoenzyme
To Be Published
|
|
7KAG
 
 | | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 3, SULFATE ION | | Authors: | Stogios, P.J, Skarina, T, Chang, C, Kim, Y, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-30 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2
To Be Published
|
|
7KES
 
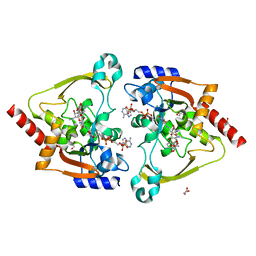 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in complex with apramycin and CoA | | Descriptor: | APRAMYCIN, Aminoglycoside N(3)-acetyltransferase, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Xu, Z, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-10-12 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
7JM1
 
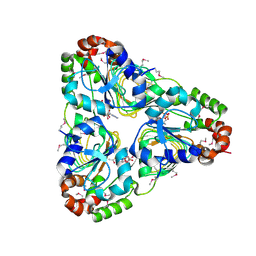 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, Aminocyclitol acetyltransferase ApmA | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-30 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with acetyl-CoA
To Be Published
|
|
7JM2
 
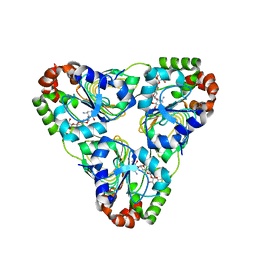 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with apramycin | | Descriptor: | APRAMYCIN, Aminocyclitol acetyltransferase ApmA, CHLORIDE ION | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-30 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with apramycin
To Be Published
|
|
7JM0
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, apoenzyme | | Descriptor: | Aminocyclitol acetyltransferase ApmA, SULFATE ION | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-30 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structure of aminoglycoside resistance enzyme ApmA, apoenzyme
To Be Published
|
|
