1LTQ
 
 | | CRYSTAL STRUCTURE OF T4 POLYNUCLEOTIDE KINASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, POLYNUCLEOTIDE KINASE | | Authors: | Galburt, E.A, Pelletier, J, Wilson, G, Stoddard, B.L. | | Deposit date: | 2002-05-20 | | Release date: | 2002-10-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure of a tRNA repair enzyme and molecular biology workhorse: T4 polynucleotide kinase.
Structure, 10, 2002
|
|
3KO2
 
 | | I-MsoI re-designed for altered DNA cleavage specificity (-7C) | | Descriptor: | 5'-D(*CP*GP*GP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*GP*TP*CP*TP*GP*C)-3', 5'-D(*GP*CP*AP*GP*AP*CP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*CP*CP*G)-3', CALCIUM ION, ... | | Authors: | Taylor, G.K, Stoddard, B.L. | | Deposit date: | 2009-11-13 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Computational reprogramming of homing endonuclease specificity at multiple adjacent base pairs.
Nucleic Acids Res., 38, 2010
|
|
6XR1
 
 | |
6XR2
 
 | |
1IQD
 
 | | Human Factor VIII C2 Domain complexed to human monoclonal BO2C11 Fab. | | Descriptor: | HUMAN FACTOR VIII, HUMAN MONOCLONAL BO2C11 FAB HEAVY CHAIN, HUMAN MONOCLONAL BO2C11 FAB LIGHT CHAIN | | Authors: | Spiegel Jr, P.C, Jacquemin, M, Saint-Remy, J.M, Stoddard, B.L, Pratt, K.P. | | Deposit date: | 2001-07-21 | | Release date: | 2001-08-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a factor VIII C2 domain-immunoglobulin G4kappa Fab complex: identification of an inhibitory antibody epitope on the surface of factor VIII.
Blood, 98, 2001
|
|
1YSD
 
 | | Yeast Cytosine Deaminase Double Mutant | | Descriptor: | CALCIUM ION, Cytosine deaminase, ZINC ION | | Authors: | Korkegian, A, Black, M.E, Baker, D, Stoddard, B.L. | | Deposit date: | 2005-02-08 | | Release date: | 2005-05-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computational thermostabilization of an enzyme.
Science, 308, 2005
|
|
1YSB
 
 | | Yeast Cytosine Deaminase Triple Mutant | | Descriptor: | CALCIUM ION, Cytosine deaminase, ZINC ION | | Authors: | Korkegian, A, Black, M.E, Baker, D, Stoddard, B.L. | | Deposit date: | 2005-02-08 | | Release date: | 2005-05-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational thermostabilization of an enzyme.
Science, 308, 2005
|
|
7UNJ
 
 | | De novo designed chlorophyll dimer protein with Zn pheophorbide a methyl ester matching geometry of purple bacterial special pair, SP1-ZnPPaM | | Descriptor: | 1,2-ETHANEDIOL, SP1-ZnPPaM designed chlorophyll dimer protein, SULFATE ION, ... | | Authors: | Kennedy, M.A, Stoddard, B.L, Ennist, N.M. | | Deposit date: | 2022-04-11 | | Release date: | 2023-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | De novo designed chlorophyll dimer protein with Zn pheophorbide a methyl ester matching geometry of purple bacterial special pair, SP1-ZnPPaM
To Be Published
|
|
7UNH
 
 | |
7UNI
 
 | | De novo designed chlorophyll dimer protein with Zn pheophorbide a methyl ester, SP2-ZnPPaM | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, SP2-ZnPPaM designed chlorophyll dimer protein, ... | | Authors: | Kennedy, M.A, Stoddard, B.L, Ennist, N.M. | | Deposit date: | 2022-04-11 | | Release date: | 2023-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | De novo designed chlorophyll dimer protein with Zn pheophorbide a methyl ester, SP2-ZnPPaM
To Be Published
|
|
3U0S
 
 | | Crystal Structure of an Enzyme Redesigned Through Multiplayer Online Gaming: CE6 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Diisopropyl-fluorophosphatase, GLYCEROL, ... | | Authors: | Bale, J.B, Shen, B.W, Stoddard, B.L. | | Deposit date: | 2011-09-29 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Increased Diels-Alderase activity through backbone remodeling guided by Foldit players.
Nat.Biotechnol., 30, 2012
|
|
6EG7
 
 | | BbvCI B2 dimer with I3C clusters | | Descriptor: | 1,2-ETHANEDIOL, 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, BbvCI endonuclease subunit 2, ... | | Authors: | Shen, B.W, Stoddard, B.L. | | Deposit date: | 2018-08-19 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure, subunit organization and behavior of the asymmetric Type IIT restriction endonuclease BbvCI.
Nucleic Acids Res., 47, 2019
|
|
1IPP
 
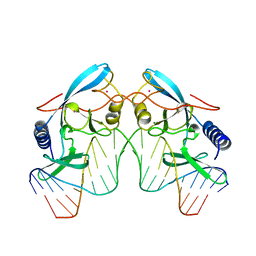 | | HOMING ENDONUCLEASE/DNA COMPLEX | | Descriptor: | CADMIUM ION, DNA (5'-D(*TP*TP*GP*AP*CP*TP*CP*TP*CP*TP*TP*AP*AP*GP*AP*GP*AP*GP*TP*CP*A)-3'), INTRON-ENCODED ENDONUCLEASE I-PPOI, ... | | Authors: | Flick, K.E, Jurica, M.S, Monnat Jr, R.J, Stoddard, B.L. | | Deposit date: | 1998-03-19 | | Release date: | 1998-09-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | DNA binding and cleavage by the nuclear intron-encoded homing endonuclease I-PpoI.
Nature, 394, 1998
|
|
6OHH
 
 | | Structure of EF1p2_mFAP2b bound to DFHBI | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, CALCIUM ION, EF1p2_mFAP2b | | Authors: | Doyle, L.A, Stoddard, B.L. | | Deposit date: | 2019-04-05 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Incorporation of sensing modalities into de novo designed fluorescence-activating proteins
Nat Commun, 12, 2021
|
|
1A73
 
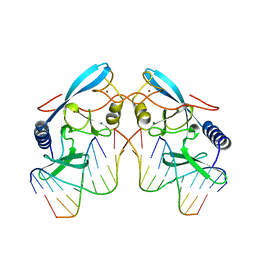 | | INTRON-ENCODED ENDONUCLEASE I-PPOI COMPLEXED WITH DNA | | Descriptor: | DNA (5'-D(*TP*TP*GP*AP*CP*TP*CP*TP*CP*TP*TP*AP*A)-3'), DNA (5'-D(P*GP*AP*GP*AP*GP*TP*CP*A)-3'), INTRON 3 (I-PPO) ENCODED ENDONUCLEASE, ... | | Authors: | Flick, K.E, Monnat Junior, R.J, Stoddard, B.L. | | Deposit date: | 1998-03-19 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DNA binding and cleavage by the nuclear intron-encoded homing endonuclease I-PpoI.
Nature, 394, 1998
|
|
299D
 
 | | CAPTURING THE STRUCTURE OF A CATALYTIC RNA INTERMEDIATE: THE HAMMERHEAD RIBOZYME | | Descriptor: | RNA HAMMERHEAD RIBOZYME | | Authors: | Scott, W.G, Murray, J.B, Arnold, J.R.P, Stoddard, B.L, Klug, A. | | Deposit date: | 1996-12-14 | | Release date: | 1997-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme.
Science, 274, 1996
|
|
2ERH
 
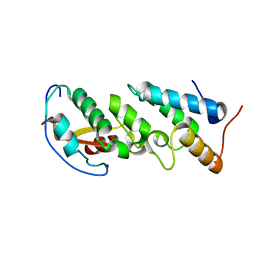 | | Crystal Structure of the E7_G/Im7_G complex; a designed interface between the colicin E7 DNAse and the Im7 immunity protein | | Descriptor: | Colicin E7, Colicin E7 immunity protein | | Authors: | Joachimiak, L.A, Kortemme, T, Stoddard, B.L, Baker, D. | | Deposit date: | 2005-10-24 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational Design of a New Hydrogen Bond Network and at Least a 300-fold Specificity Switch at a Protein-Protein Interface.
J.Mol.Biol., 361, 2006
|
|
2EX5
 
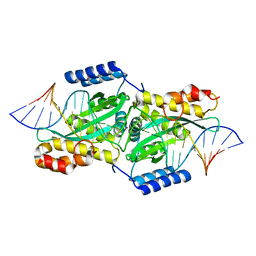 | |
2FLD
 
 | | I-MsoI Re-Designed for Altered DNA Cleavage Specificity | | Descriptor: | 5'-D(*CP*GP*GP*AP*AP*CP*GP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*CP*TP*TP*CP*TP*GP*C)-3', 5'-D(*GP*CP*AP*GP*AP*AP*GP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*CP*GP*TP*TP*CP*CP*G)-3', CALCIUM ION, ... | | Authors: | Ashworth, J, Duarte, C.M, Havranek, J.J, Sussman, D, Monnat, R.J, Stoddard, B.L, Baker, D. | | Deposit date: | 2006-01-05 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational redesign of endonuclease DNA binding and cleavage specificity.
Nature, 441, 2006
|
|
4YIT
 
 | |
4YIS
 
 | | Crystal Structure of LAGLIDADG Meganuclease I-CpaMI Bound to Uncleaved DNA | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (28-MER), ... | | Authors: | Hallinan, J.P, Kaiser, B.K, Stoddard, B.L. | | Deposit date: | 2015-03-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Indirect DNA Sequence Recognition and Its Impact on Nuclease Cleavage Activity.
Structure, 24, 2016
|
|
4YHX
 
 | |
6CZG
 
 | |
6CZJ
 
 | |
6CZH
 
 | | Structure of a redesigned beta barrel, mFAP0, bound to DFHBI | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, mFAP0 | | Authors: | Doyle, L.A, Stoddard, B.L. | | Deposit date: | 2018-04-09 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo design of a fluorescence-activating beta-barrel.
Nature, 561, 2018
|
|
