2YHS
 
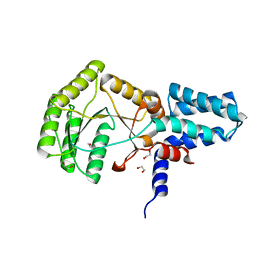 | | Structure of the E. coli SRP receptor FtsY | | Descriptor: | 1,2-ETHANEDIOL, CELL DIVISION PROTEIN FTSY | | Authors: | Stjepanovic, G, Bange, G, Wild, K, Sinning, I. | | Deposit date: | 2011-05-05 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Lipids Trigger a Conformational Switch that Regulates Signal Recognition Particle (Srp)-Mediated Protein Targeting.
J.Biol.Chem., 286, 2011
|
|
5NIY
 
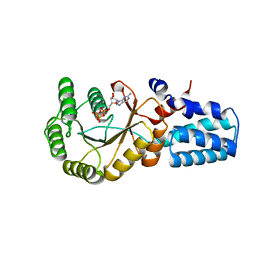 | | Signal recognition particle-docking protein FtsY | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Signal recognition particle-docking protein FtsY | | Authors: | Kempf, G, Stjepanovic, G, Lapouge, K, Sinning, I. | | Deposit date: | 2017-03-27 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Escherichia coli SRP Receptor Forms a Homodimer at the Membrane.
Structure, 26, 2018
|
|
9IVD
 
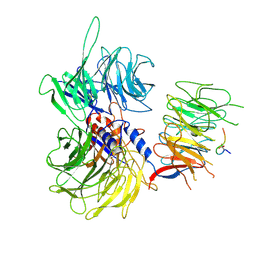 | | Cryo-EM structure of CyclinD1 bound AMBRA1-DDB1 | | Descriptor: | Activating molecule in BECN1-regulated autophagy protein 1, DNA damage-binding protein 1, G1/S-specific cyclin-D1 | | Authors: | Wang, Y, Liu, M, Su, M.-Y, Stjepanovic, G. | | Deposit date: | 2024-07-23 | | Release date: | 2025-04-23 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Mechanism of D-type cyclin recognition by the AMBRA1 E3 ligase receptor.
Sci Adv, 11, 2025
|
|
8WQR
 
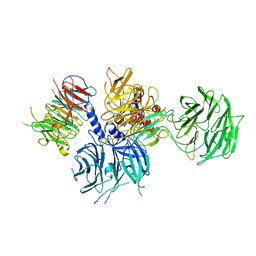 | | Structure of the DDB1-AMBRA1 E3 ligase receptor complex linked to cell cycle regulation | | Descriptor: | Activating molecule in BECN1-regulated autophagy protein 1, DNA damage-binding protein 1 | | Authors: | Liu, M, Wang, Y, Su, M.Y, Stjepanovic, G. | | Deposit date: | 2023-10-12 | | Release date: | 2023-12-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structure of the DDB1-AMBRA1 E3 ligase receptor complex linked to cell cycle regulation.
Nat Commun, 14, 2023
|
|
8Y1L
 
 | |
6C23
 
 | | Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Compact Active State | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH2, JARID2-substrate, ... | | Authors: | Kasinath, V, Faini, M, Poepsel, S, Reif, D, Feng, A, Stjepanovic, G, Aebersold, R, Nogales, E. | | Deposit date: | 2018-01-05 | | Release date: | 2018-01-24 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of human PRC2 with its cofactors AEBP2 and JARID2.
Science, 359, 2018
|
|
6C24
 
 | | Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Extended Active State | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH2, JARID2-substrate, ... | | Authors: | Kasinath, V, Faini, M, Poepsel, S, Reif, D, Feng, A, Stjepanovic, G, Aebersold, R, Nogales, E. | | Deposit date: | 2018-01-06 | | Release date: | 2018-01-24 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of human PRC2 with its cofactors AEBP2 and JARID2.
Science, 359, 2018
|
|
8KBX
 
 | | Cryo-EM structure of human ATG2A-WIPI4 complex | | Descriptor: | Autophagy-related protein 2 homolog A, WD repeat domain phosphoinositide-interacting protein 4 | | Authors: | Wang, Y, Stjepanovic, G. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-07 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis for lipid transfer by the ATG2A-ATG9A complex.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8KBY
 
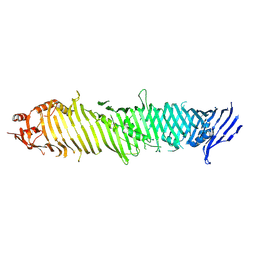 | | Cryo-EM structure of ATG2A | | Descriptor: | Autophagy-related protein 2 homolog A | | Authors: | Wang, Y, Stjepanovic, G. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-07 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis for lipid transfer by the ATG2A-ATG9A complex.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8KC3
 
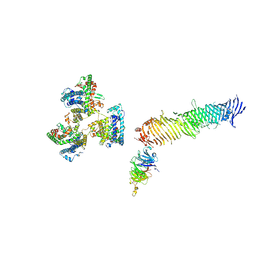 | |
3BS6
 
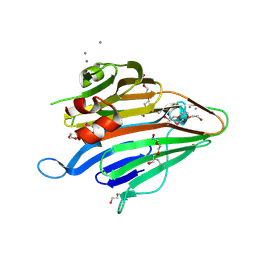 | |
3IQX
 
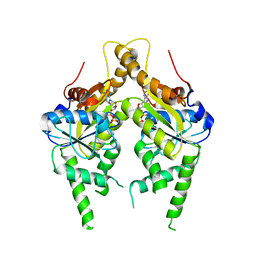 | | ADP complex of C.therm. Get3 in closed form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Tail-anchored protein targeting factor Get3, ... | | Authors: | Bozkurt, G, Wild, K, Sinning, I. | | Deposit date: | 2009-08-21 | | Release date: | 2009-12-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural insights into tail-anchored protein binding and membrane insertion by Get3.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3IQW
 
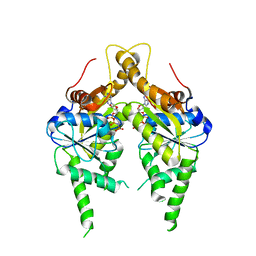 | | AMPPNP complex of C. therm. Get3 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Tail-anchored protein targeting factor Get3, ... | | Authors: | Bozkurt, G, Wild, K, Sinning, I. | | Deposit date: | 2009-08-21 | | Release date: | 2009-12-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into tail-anchored protein binding and membrane insertion by Get3.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5JW9
 
 | |
5VXV
 
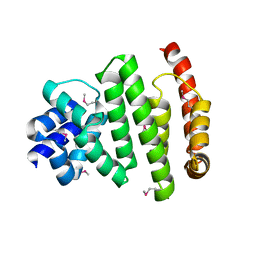 | | Peroxisomal membrane protein PEX15 | | Descriptor: | Peroxisomal membrane protein PEX15 | | Authors: | Gardner, B.M, Castanzo, D.T. | | Deposit date: | 2017-05-24 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The peroxisomal AAA-ATPase Pex1/Pex6 unfolds substrates by processive threading.
Nat Commun, 9, 2018
|
|
5L1Z
 
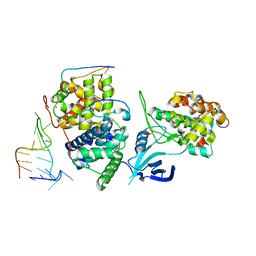 | | TAR complex with HIV-1 Tat-AFF4-P-TEFb | | Descriptor: | AF4/FMR2 family member 4, Cyclin-T1, Cyclin-dependent kinase 9, ... | | Authors: | Schulze-Gahmen, U, Hurley, J. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (5.9 Å) | | Cite: | Insights into HIV-1 proviral transcription from integrative structure and dynamics of the Tat:AFF4:P-TEFb:TAR complex.
Elife, 5, 2016
|
|
5C50
 
 | |
4GMO
 
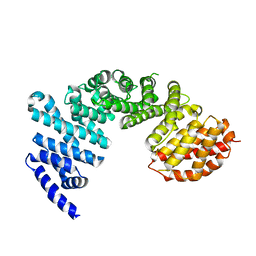 | | Crystal structure of Syo1 | | Descriptor: | Putative uncharacterized protein | | Authors: | Bange, G, Sinning, I. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synchronizing nuclear import of ribosomal proteins with ribosome assembly.
Science, 338, 2012
|
|
4GMN
 
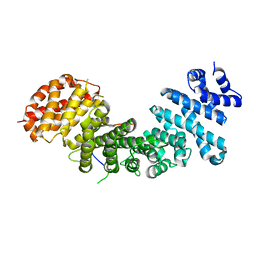 | | Structural basis of Rpl5 recognition by Syo1 | | Descriptor: | 60S ribosomal protein l5-like protein, Putative uncharacterized protein | | Authors: | Bange, G, Sinning, I. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Synchronizing nuclear import of ribosomal proteins with ribosome assembly.
Science, 338, 2012
|
|
8W4J
 
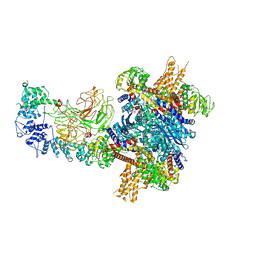 | |
8GNJ
 
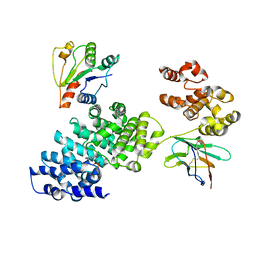 | | Human SARM1 bounded with NMN and Nanobody-C6, Conformation 2 | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NAD(+) hydrolase SARM1, Nanobody-C6 | | Authors: | Cai, Y, Zhang, H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | A conformation-specific nanobody targeting the nicotinamide mononucleotide-activated state of SARM1.
Nat Commun, 13, 2022
|
|
8GNI
 
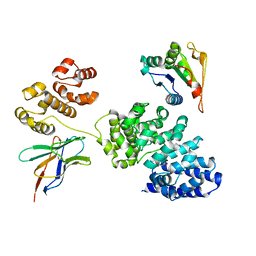 | | Human SARM1 bounded with NMN and Nanobody-C6, Conformation 1 | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NAD(+) hydrolase SARM1, Nanobody C6 | | Authors: | Cai, Y, Zhang, H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | A conformation-specific nanobody targeting the nicotinamide mononucleotide-activated state of SARM1.
Nat Commun, 13, 2022
|
|
8GQ5
 
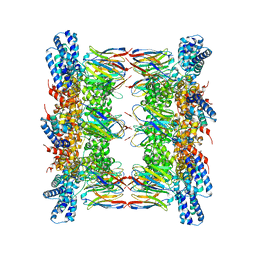 | |
6B9X
 
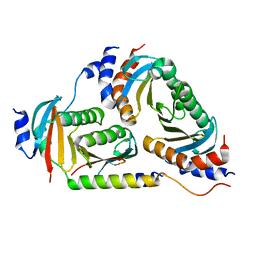 | | Crystal structure of Ragulator | | Descriptor: | Hepatitis B virus x interacting protein, Ragulator complex protein LAMTOR1, Ragulator complex protein LAMTOR2, ... | | Authors: | SU, M.-Y, Hurley, J.H. | | Deposit date: | 2017-10-11 | | Release date: | 2017-11-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Hybrid Structure of the RagA/C-Ragulator mTORC1 Activation Complex.
Mol. Cell, 68, 2017
|
|
8KBZ
 
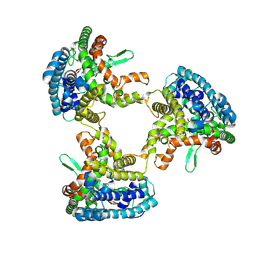 | |
