5U5O
 
 | |
5UQ0
 
 | |
5UR1
 
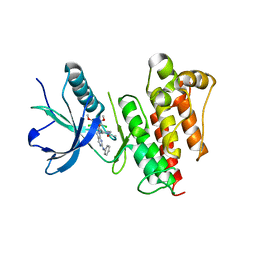 | | FGFR1 kinase domain complex with SN37333 in reversible binding mode | | Descriptor: | 3-(2,6-dichloro-3,5-dimethoxyphenyl)-1-{1-[4-(dimethylamino)but-2-enoyl]piperidin-4-yl}-7-(phenylamino)-3,4-dihydropyrimido[4,5-d]pyrimidin-2(1H)-one, Fibroblast growth factor receptor 1 | | Authors: | Yosaatmadja, Y, Paik, W.-K, Smaill, J.B, Squire, C.J. | | Deposit date: | 2017-02-09 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2-Oxo-3, 4-dihydropyrimido[4, 5-d]pyrimidinyl derivatives as new irreversible pan fibroblast growth factor receptor (FGFR) inhibitors.
Eur J Med Chem, 135, 2017
|
|
4WKQ
 
 | |
4XOO
 
 | | FMN complex of coenzyme F420:L-glutamate ligase (FbiB) from Mycobacterium tuberculosis (C-terminal domain) | | Descriptor: | Coenzyme F420:L-glutamate ligase, FLAVIN MONONUCLEOTIDE | | Authors: | Rehan, A.M, Bashiri, G, Baker, H.M, Baker, E.N, Squire, C.J. | | Deposit date: | 2015-01-16 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Elongation of the Poly-gamma-glutamate Tail of F420 Requires Both Domains of the F420: gamma-Glutamyl Ligase (FbiB) of Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
4XOQ
 
 | | F420 complex of coenzyme F420:L-glutamate ligase (FbiB) from Mycobacterium tuberculosis (C-terminal domain) | | Descriptor: | COENZYME F420, Coenzyme F420:L-glutamate ligase, SULFATE ION | | Authors: | Rehan, A.M, Bashiri, G, Baker, H.M, Baker, E.N, Squire, C.J. | | Deposit date: | 2015-01-16 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Elongation of the Poly-gamma-glutamate Tail of F420 Requires Both Domains of the F420: gamma-Glutamyl Ligase (FbiB) of Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
4XOM
 
 | | Coenzyme F420:L-glutamate ligase (FbiB) from Mycobacterium tuberculosis (C-terminal domain). | | Descriptor: | Coenzyme F420:L-glutamate ligase, SULFATE ION | | Authors: | Rehan, A.M, Bashiri, G, Baker, H.M, Baker, E.N, Squire, C.J. | | Deposit date: | 2015-01-16 | | Release date: | 2016-02-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Elongation of the Poly-gamma-glutamate Tail of F420 Requires Both Domains of the F420: gamma-Glutamyl Ligase (FbiB) of Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
6ANN
 
 | |
6ANM
 
 | |
6C19
 
 | | FGFR1 kinase complex with inhibitor SN36985 | | Descriptor: | 3-(2,6-dichloro-3,5-dimethoxyphenyl)-1-{(3S)-1-[(2E)-4-(dimethylamino)but-2-enoyl]pyrrolidin-3-yl}-7-(methylamino)-3,4-dihydropyrimido[4,5-d]pyrimidin-2(1H)-one, Fibroblast growth factor receptor 1, SULFATE ION | | Authors: | Yosaatmadja, Y, Smaill, J.B, Squire, C.J. | | Deposit date: | 2018-01-04 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Understanding the structural requirements for covalent inhibition of FGFR1-3
To Be Published
|
|
6C1O
 
 | | FGFR1 kinase domain complexed with FIIN-1 | | Descriptor: | Fibroblast growth factor receptor 1, N-(3-{[3-(2,6-dichloro-3,5-dimethoxyphenyl)-7-{[4-(diethylamino)butyl]amino}-2-oxo-3,4-dihydropyrimido[4,5-d]pyrimidin-1(2H)-yl]methyl}phenyl)prop-2-enamide, SULFATE ION | | Authors: | Kalyukina, M, Yosaatmadja, Y, Smaill, J.B, Squire, C.J. | | Deposit date: | 2018-01-05 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A new class of FGFR1 inhibitors
To Be Published
|
|
6C1B
 
 | | FGFR1 kinase complex with inhibitor SN37118 | | Descriptor: | 3-(2,6-dichloro-3,5-dimethoxyphenyl)-1-{(3S)-1-[(2E)-4-(dimethylamino)but-2-enoyl]pyrrolidin-3-yl}-7-(phenylamino)-3,4-dihydropyrimido[4,5-d]pyrimidin-2(1H)-one, Fibroblast growth factor receptor 1 | | Authors: | Yosaatmadja, Y, Smaill, J.B, Squire, C.J. | | Deposit date: | 2018-01-04 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Understanding the structural requirements for covalent inhibition of FGFR1-3
To Be Published
|
|
6C18
 
 | | FGFR1 kinase complex with inhibitor SN37115 | | Descriptor: | 3-(2,6-dichloro-3,5-dimethoxyphenyl)-1-{(3S)-1-[(2E)-4-(dimethylamino)but-2-enoyl]pyrrolidin-3-yl}-7-[(propan-2-yl)amino]-3,4-dihydropyrimido[4,5-d]pyrimidin-2(1H)-one, Fibroblast growth factor receptor 1, SULFATE ION | | Authors: | Yosaatmadja, Y, Smaill, J.B, Squire, C.J. | | Deposit date: | 2018-01-04 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Understanding the structural requirements for covalent inhibition of FGFR1-3
To Be Published
|
|
2O7V
 
 | | Carboxylesterase AeCXE1 from Actinidia eriantha covalently inhibited by paraoxon | | Descriptor: | CXE carboxylesterase, DIETHYL PHOSPHONATE | | Authors: | Ileperuma, N.R, Marshall, S.D, Squire, C.J, Baker, H.M, Oakeshott, J.G, Russell, R.J, Plummer, K.M, Newcomb, R.D, Baker, E.N. | | Deposit date: | 2006-12-11 | | Release date: | 2007-02-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High-Resolution Crystal Structure of Plant Carboxylesterase AeCXE1, from Actinidia eriantha, and Its Complex with a High-Affinity Inhibitor Paraoxon.
Biochemistry, 46, 2007
|
|
2O7R
 
 | | Plant carboxylesterase AeCXE1 from Actinidia eriantha with acyl adduct | | Descriptor: | CXE carboxylesterase, PROPYL ACETATE | | Authors: | Ileperuma, N.R, Marshall, S.D, Squire, C.J, Baker, H.M, Oakeshott, J.G, Russell, R.J, Plummer, K.M, Newcomb, R.D, Baker, E.N. | | Deposit date: | 2006-12-11 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-Resolution Crystal Structure of Plant Carboxylesterase AeCXE1, from Actinidia eriantha, and Its Complex with a High-Affinity Inhibitor Paraoxon.
Biochemistry, 46, 2007
|
|
6MW0
 
 | | Mle-Phe-Mle-D-Phe. Linear tetrapeptide related to pseudoxylallemycin A. | | Descriptor: | METHANOL, Mle-Phe-Mle-D-Phe Linear tetrapeptide related to pseudoxylallemycin A | | Authors: | Cameron, A.J, Harris, P.W.R, Brimble, M.A, Squire, C.J. | | Deposit date: | 2018-10-29 | | Release date: | 2019-09-11 | | Method: | X-RAY DIFFRACTION (0.78 Å) | | Cite: | Investigations of the key macrolactamisation step in the synthesis of cyclic tetrapeptide pseudoxylallemycin A.
Org.Biomol.Chem., 17, 2019
|
|
6MW1
 
 | |
6MZQ
 
 | | TAS-120 in reversible binding mode with FGFR1 | | Descriptor: | 1-[(3S)-3-{4-amino-3-[(3,5-dimethoxyphenyl)ethynyl]-1H-pyrazolo[3,4-d]pyrimidin-1-yl}pyrrolidin-1-yl]prop-2-en-1-one, Fibroblast growth factor receptor 1 | | Authors: | Kalyukina, M, Squire, C.J. | | Deposit date: | 2018-11-05 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | TAS-120 Cancer Target Binding: Defining Reactivity and Revealing the First Fibroblast Growth Factor Receptor 1 (FGFR1) Irreversible Structure.
ChemMedChem, 14, 2019
|
|
6MZW
 
 | | TAS-120 covalent complex with FGFR1 | | Descriptor: | 1-[(3S)-3-{4-amino-3-[(3,5-dimethoxyphenyl)ethynyl]-1H-pyrazolo[3,4-d]pyrimidin-1-yl}pyrrolidin-1-yl]prop-2-en-1-one, Fibroblast growth factor receptor 1, SULFATE ION | | Authors: | Kalyukina, M, Squire, C.J. | | Deposit date: | 2018-11-05 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | TAS-120 Cancer Target Binding: Defining Reactivity and Revealing the First Fibroblast Growth Factor Receptor 1 (FGFR1) Irreversible Structure.
ChemMedChem, 14, 2019
|
|
6MW2
 
 | |
6MVZ
 
 | | Mle-Phe-Mle-Phe. Linear precursor of pseudoxylallemycin A. | | Descriptor: | Linear precursor of pseudoxylallemycin A, trifluoroacetic acid | | Authors: | Cameron, A.J, Harris, P.W.R, Brimble, M.A, Squire, C.J. | | Deposit date: | 2018-10-29 | | Release date: | 2019-09-11 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | Investigations of the key macrolactamisation step in the synthesis of cyclic tetrapeptide pseudoxylallemycin A.
Org.Biomol.Chem., 17, 2019
|
|
6NVJ
 
 | | FGFR4 complex with N-(2-((5-((2,6-dichloro-3,5-dimethoxybenzyl)oxy)pyrimidin-2-yl)amino)-3-fluorophenyl)acrylamide | | Descriptor: | Fibroblast growth factor receptor 4, N-[2-({5-[(2,6-dichloro-3,5-dimethoxyphenyl)methoxy]pyrimidin-2-yl}amino)-3-fluorophenyl]propanamide, SULFATE ION | | Authors: | Lin, X, Smaill, J.B, Squire, C.J, Yosaatmadja, Y. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rotational Freedom, Steric Hindrance, and Protein Dynamics Explain BLU554 Selectivity for the Hinge Cysteine of FGFR4.
Acs Med.Chem.Lett., 10, 2019
|
|
6NVH
 
 | | FGFR4 complex with N-(2-((5-((2,6-dichloro-3,5-dimethoxybenzyl)oxy)pyrimidin-2-yl)amino)-3-methylphenyl)acrylamide | | Descriptor: | Fibroblast growth factor receptor 4, N-[2-({5-[(2,6-dichloro-3,5-dimethoxyphenyl)methoxy]pyrimidin-2-yl}amino)-3-methylphenyl]propanamide, SULFATE ION | | Authors: | Lin, X, Smaill, J.B, Squire, C.J. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rotational Freedom, Steric Hindrance, and Protein Dynamics Explain BLU554 Selectivity for the Hinge Cysteine of FGFR4.
Acs Med.Chem.Lett., 10, 2019
|
|
6NVL
 
 | | FGFR1 complex with N-(2-((5-((2,6-dichloro-3,5-dimethoxybenzyl)oxy)pyrimidin-2-yl)amino)-3-methylphenyl)acrylamide | | Descriptor: | Fibroblast growth factor receptor 1, N-[2-({5-[(2,6-dichloro-3,5-dimethoxyphenyl)methoxy]pyrimidin-2-yl}amino)-3-methylphenyl]propanamide, SULFATE ION | | Authors: | Lin, X, Smaill, J.B, Squire, C.J. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Rotational Freedom, Steric Hindrance, and Protein Dynamics Explain BLU554 Selectivity for the Hinge Cysteine of FGFR4.
Acs Med.Chem.Lett., 10, 2019
|
|
6NVG
 
 | | FGFR4 complex with N-(3,5-dichloro-2-((5-((2,6-dichloro-3,5-dimethoxybenzyl)oxy)pyrimidin-2-yl)amino)phenyl)acrylamide | | Descriptor: | Fibroblast growth factor receptor 4, N-[3,5-dichloro-2-({5-[(2,6-dichloro-3,5-dimethoxyphenyl)methoxy]pyrimidin-2-yl}amino)phenyl]propanamide, SULFATE ION | | Authors: | Lin, X, Smaill, J.B, Squire, C.J, Yosaatmadja, Y. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Rotational Freedom, Steric Hindrance, and Protein Dynamics Explain BLU554 Selectivity for the Hinge Cysteine of FGFR4.
Acs Med.Chem.Lett., 10, 2019
|
|
