1GIL
 
 | |
1GFI
 
 | | STRUCTURES OF ACTIVE CONFORMATIONS OF GI ALPHA 1 AND THE MECHANISM OF GTP HYDROLYSIS | | Descriptor: | GUANINE NUCLEOTIDE-BINDING PROTEIN G, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Coleman, D.E, Berghuis, A.M, Sprang, S.R. | | Deposit date: | 1994-11-11 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of active conformations of Gi alpha 1 and the mechanism of GTP hydrolysis.
Science, 265, 1994
|
|
1D2Z
 
 | | THREE-DIMENSIONAL STRUCTURE OF A COMPLEX BETWEEN THE DEATH DOMAINS OF PELLE AND TUBE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DEATH DOMAIN OF PELLE, DEATH DOMAIN OF TUBE | | Authors: | Xiao, T, Towb, P, Wasserman, S.A, Sprang, S.R. | | Deposit date: | 1999-09-28 | | Release date: | 1999-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of a complex between the death domains of Pelle and Tube.
Cell(Cambridge,Mass.), 99, 1999
|
|
1CUL
 
 | | COMPLEX OF GS-ALPHA WITH THE CATALYTIC DOMAINS OF MAMMALIAN ADENYLYL CYCLASE: COMPLEX WITH 2',5'-DIDEOXY-ADENOSINE 3'-TRIPHOSPHATE AND MG | | Descriptor: | 2',5'-DIDEOXY-ADENOSINE 3'-MONOPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ... | | Authors: | Tesmer, J.J.G, Dessauer, C.A, Sunahara, R.K, Johnson, R.A, Gilman, A.G, Sprang, S.R. | | Deposit date: | 1999-08-20 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for P-site inhibition of adenylyl cyclase.
Biochemistry, 39, 2000
|
|
1GG2
 
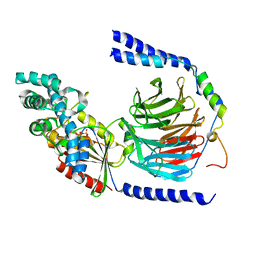 | |
1GP2
 
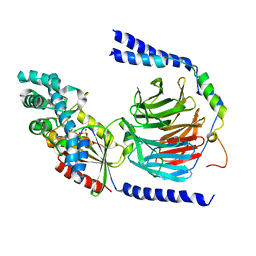 | |
1PYG
 
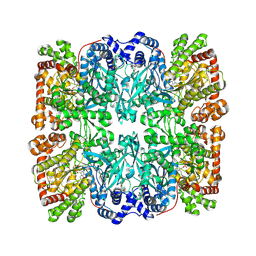 | |
5KKE
 
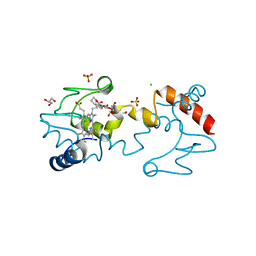 | |
5KLU
 
 | |
4WEE
 
 | |
9GPB
 
 | |
1CIP
 
 | |
8GPB
 
 | |
7GPB
 
 | | STRUCTURAL MECHANISM FOR GLYCOGEN PHOSPHORYLASE CONTROL BY PHOSPHORYLATION AND AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCOGEN PHOSPHORYLASE B, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Barford, D, Hu, S.-H, Johnson, L.N. | | Deposit date: | 1990-11-13 | | Release date: | 1992-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural mechanism for glycogen phosphorylase control by phosphorylation and AMP.
J.Mol.Biol., 218, 1991
|
|
4TSV
 
 | |
7RLE
 
 | |
7TGP
 
 | | E176T/Y188G variant of the internal UBA Domain of HHR23A | | Descriptor: | UV excision repair protein RAD23 homolog A | | Authors: | Rothfuss, M, Bowler, B.E, McClelland, L.J, Sprang, S.R. | | Deposit date: | 2022-01-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-Accuracy Prediction of Stabilizing Surface Mutations to the Three-Helix Bundle, UBA(1), with EmCAST.
J.Am.Chem.Soc., 145, 2023
|
|
4GPB
 
 | |
1A0R
 
 | | HETEROTRIMERIC COMPLEX OF PHOSDUCIN/TRANSDUCIN BETA-GAMMA | | Descriptor: | FARNESYL, PHOSDUCIN, TRANSDUCIN (BETA SUBUNIT), ... | | Authors: | Loew, A, Ho, Y.-K, Blundell, T.L, Bax, B. | | Deposit date: | 1997-12-05 | | Release date: | 1998-12-30 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phosducin induces a structural change in transducin beta gamma.
Structure, 6, 1998
|
|
1EDY
 
 | |
6ODL
 
 | | Crystal structure of GluN2A agonist binding domain with 4-butyl-(S)-CCG-IV | | Descriptor: | (1S,2R)-2-[(S)-amino(carboxy)methyl]-1-butylcyclopropane-1-carboxylic acid, Glutamate receptor ionotropic, NMDA 2A,Glutamate receptor ionotropic, ... | | Authors: | Mou, T.C, Clausen, R.P, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Stereoselective synthesis of novel 2'-(S)-CCG-IV analogues as potent NMDA receptor agonists.
Eur.J.Med.Chem., 212, 2021
|
|
6D94
 
 | | Crystal structure of PPAR gamma in complex with Mediator of RNA polymerase II transcription subunit 1 | | Descriptor: | (2~{S})-3-[4-[2-[methyl(pyridin-2-yl)amino]ethoxy]phenyl]-2-[[2-(phenylcarbonyl)phenyl]amino]propanoic acid, Mediator of RNA polymerase II transcription subunit 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Mou, T.C, Chrisman, I.M, Hughes, T.S, Sprang, S.R. | | Deposit date: | 2018-04-27 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structural mechanism of nuclear receptor biased agonism.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6GPB
 
 | |
3GPB
 
 | |
2TNF
 
 | | 1.4 A RESOLUTION STRUCTURE OF MOUSE TUMOR NECROSIS FACTOR, TOWARDS MODULATION OF ITS SELECTIVITY AND TRIMERISATION | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ISOPROPYL ALCOHOL, PROTEIN (TUMOR NECROSIS FACTOR ALPHA) | | Authors: | Baeyens, K.J, De Bondt, H.L, Raeymaekers, A, Fiers, W, De Ranter, C.J. | | Deposit date: | 1998-10-12 | | Release date: | 1999-10-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structure of mouse tumour-necrosis factor at 1.4 A resolution: towards modulation of its selectivity and trimerization.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
