4URP
 
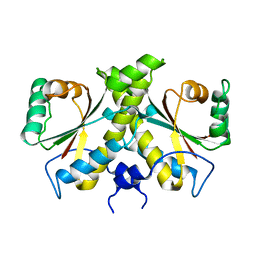 | | The Crystal structure of Nitroreductase from Saccharomyces cerevisiae | | Descriptor: | FATTY ACID REPRESSION MUTANT PROTEIN 2 | | Authors: | Song, H.-N, Woo, E.-J, Bang, S.-Y, Jung, D.-G, Park, S.-G. | | Deposit date: | 2014-07-01 | | Release date: | 2015-04-29 | | Last modified: | 2015-07-08 | | Method: | X-RAY DIFFRACTION (2.991 Å) | | Cite: | Crystal Structure of the Fungal Nitroreductase Frm2 from Saccharomyces Cerevisiae.
Protein Sci., 24, 2015
|
|
4QII
 
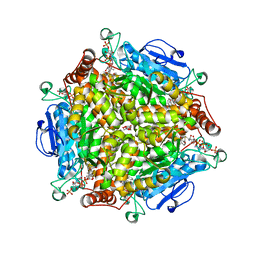 | | Crystal Structure of type II MenB from Mycobacteria tuberculosis | | Descriptor: | 1,4-Dihydroxy-2-naphthoyl-CoA synthase, Salicylyl CoA, TRIETHYLENE GLYCOL | | Authors: | Song, H.G, Tse, Y.S, Sung, H.P, Guo, Z.H. | | Deposit date: | 2014-05-31 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Ligand-dependent active-site closure revealed in the crystal structure of Mycobacterium tuberculosis MenB complexed with product analogues
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4QIJ
 
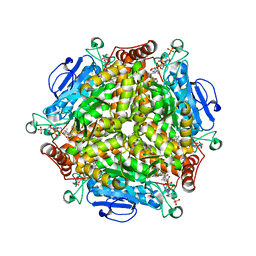 | | Crystal structure of MenB from Mycobacteria tuberculosis in complex with 1-HNA-CoA | | Descriptor: | 1,4-Dihydroxy-2-naphthoyl-CoA synthase, 1-hydroxy-2-naphthoyl-CoA | | Authors: | Song, H.G, Sung, H.P, Tse, Y.S, Guo, Z.H. | | Deposit date: | 2014-05-31 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ligand-dependent active-site closure revealed in the crystal structure of Mycobacterium tuberculosis MenB complexed with product analogues
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4U1T
 
 | |
4U2H
 
 | |
7V97
 
 | | Arsenic-bound p53 DNA-binding domain mutant V272M | | Descriptor: | ARSENIC, Cellular tumor antigen p53, ZINC ION | | Authors: | Lu, M, Xing, Y.F, Wang, Z.Y, Ni, Y, Song, H.X. | | Deposit date: | 2021-08-24 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Diverse rescue potencies of p53 mutations to ATO are predetermined by intrinsic mutational properties.
Sci Transl Med, 15, 2023
|
|
1DT9
 
 | |
1LZR
 
 | |
6XNG
 
 | | MHC-like protein complex structure | | Descriptor: | (3R)-N-[(2S,3R)-1-(alpha-D-galactopyranosyloxy)-3-hydroxyheptadecan-2-yl]-3-hydroxyheptadecanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Thirunavukkarasu, P, Le Nours, J, Rossjohn, J. | | Deposit date: | 2020-07-02 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Host immunomodulatory lipids created by symbionts from dietary amino acids.
Nature, 600, 2021
|
|
3ZHL
 
 | |
2VR5
 
 | | Crystal structure of Trex from Sulfolobus Solfataricus in complex with acarbose intermediate and glucose | | Descriptor: | 4-O-(4,6-dideoxy-4-{[(1S,2S,3S,4R,5S)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl]amino}-alpha-D-glucopyranosyl)-beta-D-glucopyranose, GLYCEROL, GLYCOGEN OPERON PROTEIN GLGX, ... | | Authors: | Song, H.-N, Yoon, S.-M, Lee, S.-J, Cha, H.-J, Park, K.-H, Woo, E.-J. | | Deposit date: | 2008-03-26 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insight Into the Bifunctional Mechanism of the Glycogen-Debranching Enzyme Trex from the Archaeon Sulfolobus Solfataricus.
J.Biol.Chem., 283, 2008
|
|
2VUY
 
 | | Crystal structure of Glycogen Debranching exzyme TreX from Sulfolobus solfatarius | | Descriptor: | GLYCOGEN OPERON PROTEIN GLGX | | Authors: | Song, H.-N, Yoon, S.-M, Cha, H.-J, Park, K.-H, Woo, E.-J. | | Deposit date: | 2008-06-02 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insight Into the Bifunctional Mechanism of the Glycogen-Debranching Enzyme Trex from the Archaeon Sulfolobus Solfataricus.
J.Biol.Chem., 283, 2008
|
|
5JHL
 
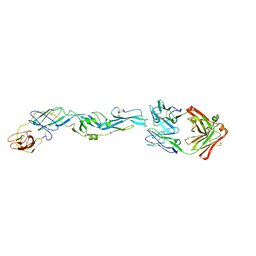 | | Crystal structure of zika virus envelope protein in complex with a flavivirus broadly-protective antibody | | Descriptor: | Antibody Heavy chain, antibody Light chain, envelope protein | | Authors: | Dai, L, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2016-04-21 | | Release date: | 2016-05-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the Zika Virus Envelope Protein and Its Complex with a Flavivirus Broadly Protective Antibody.
Cell Host Microbe, 19, 2016
|
|
5JHM
 
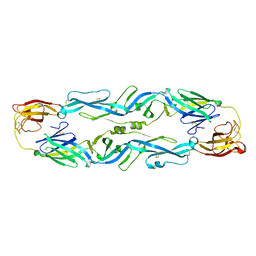 | |
4Y5T
 
 | | Structure of FtmOx1 apo with metal Iron | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COBALT (II) ION, FE (II) ION, ... | | Authors: | Yan, W, Zhang, Y. | | Deposit date: | 2015-02-12 | | Release date: | 2015-11-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Endoperoxide formation by an alpha-ketoglutarate-dependent mononuclear non-haem iron enzyme.
Nature, 527, 2015
|
|
4Y5S
 
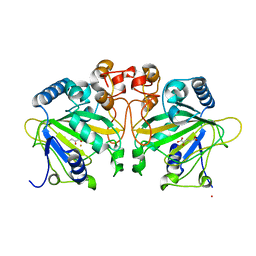 | | Structure of FtmOx1 with a-Ketoglutarate as co-substrate | | Descriptor: | 2-OXOGLUTARIC ACID, COBALT (II) ION, FE (II) ION, ... | | Authors: | Yan, W, Zhang, Y. | | Deposit date: | 2015-02-12 | | Release date: | 2015-11-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.543 Å) | | Cite: | Endoperoxide formation by an alpha-ketoglutarate-dependent mononuclear non-haem iron enzyme.
Nature, 527, 2015
|
|
6EF4
 
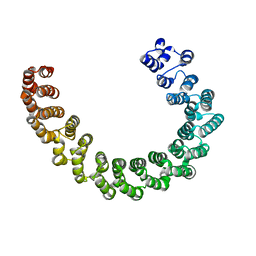 | | Crystal structure of mouse PP2A Aalpha P179R mutant | | Descriptor: | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform | | Authors: | Wang, Z, Shen, G, Xu, W. | | Deposit date: | 2018-08-16 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Highly Recurrent PP2A A alpha-Subunit Mutation P179R Alters Protein Structure and Impairs PP2A Enzyme Function to Promote Endometrial Tumorigenesis.
Cancer Res., 79, 2019
|
|
2H5E
 
 | | Crystal structure of E.coli polypeptide release factor RF3 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Peptide chain release factor RF-3 | | Authors: | Song, H.W, Zhou, Z.H. | | Deposit date: | 2006-05-26 | | Release date: | 2007-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors
Cell(Cambridge,Mass.), 129, 2007
|
|
3ZHD
 
 | |
3ZHK
 
 | |
2GIZ
 
 | | Structural and functional analysis of Natrin, a member of crisp-3 family blocks a variety of ion channels | | Descriptor: | Natrin-1 | | Authors: | Jiang, T, Wang, F, Li, H, Yin, C, Zhou, Y, Shu, Y, Qi, Z, Lin, Z. | | Deposit date: | 2006-03-30 | | Release date: | 2006-11-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and functional analysis of natrin, a venom protein that targets various ion channels
Biochem.Biophys.Res.Commun., 351, 2006
|
|
6J6M
 
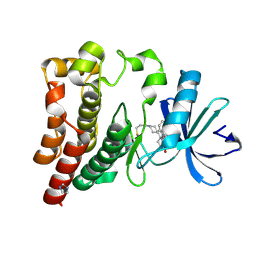 | | Co-crystal structure of BTK kinase domain with Zanubrutinib | | Descriptor: | (7S)-2-(4-phenoxyphenyl)-7-(1-propanoylpiperidin-4-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, IMIDAZOLE, Tyrosine-protein kinase BTK | | Authors: | Zhou, X, Hong, Y. | | Deposit date: | 2019-01-15 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery of Zanubrutinib (BGB-3111), a Novel, Potent, and Selective Covalent Inhibitor of Bruton's Tyrosine Kinase.
J.Med.Chem., 62, 2019
|
|
4XG8
 
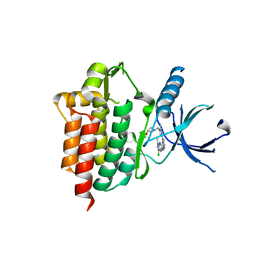 | | Crystal structure of an inhibitor-bound Syk | | Descriptor: | 1-[(1-{2-[(3-chloro-1-methyl-1H-indazol-5-yl)amino]pyrimidin-4-yl}-3-methyl-1H-pyrazol-4-yl)methyl]azetidin-3-ol, Tyrosine-protein kinase SYK | | Authors: | Lee, S.J, Choi, J, Han, B.G, Song, H, Koh, J.S, Lee, B.I. | | Deposit date: | 2014-12-30 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of spleen tyrosine kinase in complex with novel inhibitors: structural insights for design of anticancer drugs
Febs J., 283, 2016
|
|
4XG2
 
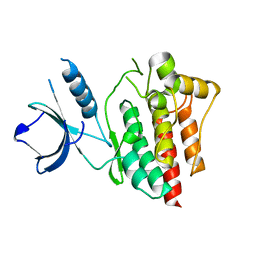 | | Crystal structure of ligand-free Syk | | Descriptor: | Tyrosine-protein kinase SYK | | Authors: | Lee, S.J, Choi, J, Han, B.G, Song, H, Koh, J.S, Lee, B.I. | | Deposit date: | 2014-12-30 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structures of spleen tyrosine kinase in complex with novel inhibitors: structural insights for design of anticancer drugs
Febs J., 283, 2016
|
|
4XG3
 
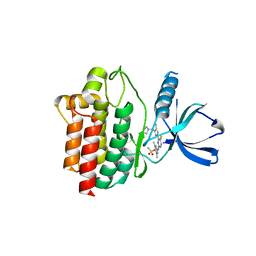 | | Crystal structure of an inhibitor-bound Syk | | Descriptor: | 4-{[5-fluoro-4-(3-{[(3R)-3-hydroxypyrrolidin-1-yl]methyl}-4-methyl-1H-pyrrol-1-yl)pyrimidin-2-yl]amino}-2,6-dimethylphenyl methanesulfonate, Tyrosine-protein kinase SYK | | Authors: | Lee, S.J, Choi, J, Han, B.G, Song, H, Koh, J.S, Lee, B.I. | | Deposit date: | 2014-12-30 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of spleen tyrosine kinase in complex with novel inhibitors: structural insights for design of anticancer drugs
Febs J., 283, 2016
|
|
