1ML8
 
 | | structural genomics | | Descriptor: | hypothetical protein (crp region) | | Authors: | Korolev, S, Skarina, T, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-08-30 | | Release date: | 2003-04-22 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural genomics
To be Published
|
|
1SFX
 
 | | X-ray crystal structure of putative HTH transcription regulator from Archaeoglobus fulgidus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Conserved hypothetical protein AF2008 | | Authors: | Osipiuk, J, Skarina, T, Savchenko, A, Edwards, A, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-20 | | Release date: | 2004-08-03 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray crystal structure of putative HTH transcription regulator from Archaeoglobus fulgidus
To be Published
|
|
5T1P
 
 | | Crystal structure of the putative periplasmic solute-binding protein from Campylobacter jejuni | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ABC transporter, ... | | Authors: | Filippova, E.V, Wawrzsak, Z, Sandoval, J, Skarina, T, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-19 | | Release date: | 2016-09-07 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the putative periplasmic solute-binding protein from Campylobacter jejuni
To Be Published
|
|
5TPM
 
 | | 2.8 Angstrom Crystal Structure of the C-terminal Dimerization Domain of Transcriptional Regulator PdhR from Escherichia coli. | | Descriptor: | Pyruvate dehydrogenase complex repressor | | Authors: | Minasov, G, Wawrzak, Z, Sandoval, J, Skarina, T, Grimshaw, S, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2.8 Angstrom Crystal Structure of the C-terminal Dimerization Domain of Transcriptional Regulator PdhR from Escherichia coli.
To Be Published
|
|
5U08
 
 | | Crystal structure of an aminoglycoside acetyltransferase meta-AAC0020 from an uncultured soil metagenomic sample in complex with sisomicin | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, ACETATE ION, CALCIUM ION, ... | | Authors: | Xu, Z, Skarina, T, Wawrzak, Z, Stogios, P.J, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-23 | | Release date: | 2017-02-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural and Functional Survey of Environmental Aminoglycoside Acetyltransferases Reveals Functionality of Resistance Enzymes.
ACS Infect Dis, 3, 2017
|
|
5UXD
 
 | | Crystal structure of macrolide 2'-phosphotransferase MphH from Brachybacterium faecium in complex with azithromycin | | Descriptor: | AZITHROMYCIN, CHLORIDE ION, Macrolide 2'-phosphotransferase MphH, ... | | Authors: | Stogios, P.J, Skarina, T, Wawrzak, Z, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
5UXC
 
 | | Crystal structure of macrolide 2'-phosphotransferase MphH from Brachybacterium faecium in complex with GDP | | Descriptor: | AZITHROMYCIN, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Wawrzak, Z, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-08-16 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
5UXB
 
 | | Crystal structure of macrolide 2'-phosphotransferase MphH from Brachybacterium faecium, apoenzyme | | Descriptor: | CHLORIDE ION, Macrolide 2'-phosphotransferase MphH | | Authors: | Stogios, P.J, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
5VIS
 
 | | 1.73 Angstrom Resolution Crystal Structure of Dihydropteroate Synthase (folP-SMZ_B27) from Soil Uncultured Bacterium. | | Descriptor: | CHLORIDE ION, D(-)-TARTARIC ACID, Dihydropteroate Synthase, ... | | Authors: | Minasov, G, Wawrzak, Z, Di Leo, R, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-17 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | 1.73 Angstrom Resolution Crystal Structure of Dihydropteroate Synthase (folP-SMZ_B27) from Soil
Uncultured Bacterium.
To Be Published
|
|
5VGC
 
 | | Crystal structure of the NleG5-1 effector (C200A) from Escherichia coli O157:H7 str. Sakai | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Borek, D, Valleau, D, Skarina, T, Jobin, M.C, Wawrzak, Z, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-10 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the NleG5-1 effector (C200A) from Escherichia coli O157:H7 str. Sakai
To Be Published
|
|
5VRV
 
 | | 2.05 Angstrom Resolution Crystal Structure of C-terminal Domain (DUF2156) of Putative Lysylphosphatidylglycerol Synthetase from Agrobacterium fabrum. | | Descriptor: | GLYCEROL, Protein regulated by acid pH, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-05-11 | | Release date: | 2017-05-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2.05 Angstrom Resolution Crystal Structure of C-terminal Domain (DUF2156) of Putative Lysylphosphatidylglycerol Synthetase from Agrobacterium fabrum.
To Be Published
|
|
3IB3
 
 | | Crystal Structure of SACOL2612 - CocE/NonD family hydrolase from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, CocE/NonD family hydrolase, NICKEL (II) ION, ... | | Authors: | Domagalski, M.J, Chruszcz, M, Osinski, T, Skarina, T, Onopriyenko, O, Cymborowski, M, Shumilin, I.A, Savchenko, A, Edwards, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-15 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of SACOL2612 - CocE/NonD family hydrolase from Staphylococcus aureus
To be Published
|
|
3III
 
 | | 1.95 Angstrom Crystal Structure of CocE/NonD family hydrolase (SACOL2612) from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, CocE/NonD family hydrolase, NICKEL (II) ION, ... | | Authors: | Osinski, T, Chruszcz, M, Domagalski, M.J, Cymborowski, M, Shumilin, I.A, Skarina, T, Onopriyenko, O, Zimmerman, M.D, Savchenko, A, Edwards, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-01 | | Release date: | 2009-08-18 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of CocE/NonD family hydrolase (SACOL2612) from Staphylococcus aureus
To be Published
|
|
1YLQ
 
 | | Crystal structure of putative nucleotidyltransferase | | Descriptor: | SULFATE ION, putative nucleotidyltransferase, hypothetical protein AF0614 | | Authors: | Chang, C, Joachimiak, A, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-01-19 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.016 Å) | | Cite: | Crystal structure of Hypothetical protein AF0614, putative nucleotidyltransferase
To be Published
|
|
1YQE
 
 | | Crystal Structure of Conserved Protein of Unknown Function AF0625 | | Descriptor: | Hypothetical UPF0204 protein AF0625, PYROPHOSPHATE 2- | | Authors: | Liu, Y, Skarina, T, Dong, A, Kudritskam, M, Savchenko, A, Pai, E.F, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-01 | | Release date: | 2005-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of Conserved Hypothetical Protein AF0625
To be Published
|
|
1YSQ
 
 | | The crystal structure of transcriptional regulator YaiJ | | Descriptor: | HTH-type transcriptional regulator yiaJ, PHOSPHATE ION | | Authors: | Bochkarev, A, Lunin, V.V, Ezersky, A, Evdokimova, E, Skarina, T, Xu, X, Borek, D, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-08 | | Release date: | 2005-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural study of effector binding specificity in IclR transcriptional regulators
To be Published
|
|
1YSP
 
 | | Crystal structure of the C-terminal domain of E. coli transcriptional regulator KdgR. | | Descriptor: | SULFATE ION, Transcriptional regulator kdgR | | Authors: | Bochkarev, A, Lunin, V.V, Ezersky, A, Evdokimova, E, Skarina, T, Xu, X, Borek, D, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-08 | | Release date: | 2005-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural study of effector binding specificity in IclR transcriptional regulators
To be Published
|
|
1ZKE
 
 | | 1.6 A Crystal Structure of a Protein HP1531 of Unknown Function from Helicobacter pylori | | Descriptor: | Hypothetical protein HP1531, MAGNESIUM ION | | Authors: | Zhang, R, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.6A crystal structure of a hypothetical protein HP1531 from Helicobacter pylori 26695
To be Published
|
|
1Z7A
 
 | | Crystal structure of probable Polysaccharide deacetylase from Pseudomonas aeruginosa PAO1 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Chang, C, Skarina, T, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-03-24 | | Release date: | 2005-05-10 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structure of probable Polysaccharide deacetylase from Pseudomonas aeruginosa PAO1
To be Published
|
|
1ZX5
 
 | | The structure of a putative mannosephosphate isomerase from Archaeoglobus fulgidus | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, GLYCEROL, ... | | Authors: | Cuff, M.E, Skarina, T, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-06-06 | | Release date: | 2005-07-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of a putative mannosephosphate isomerase from Archaeoglobus fulgidus
To be Published
|
|
3BIG
 
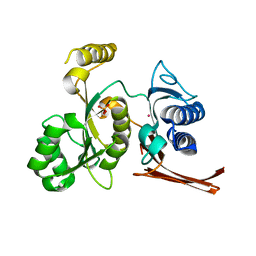 | | Crystal structure of the fructose-1,6-bisphosphatase GlpX from E.coli in complex with inorganic phosphate | | Descriptor: | Fructose-1,6-bisphosphatase class II glpX, PHOSPHATE ION, UNKNOWN ATOM OR ION | | Authors: | Lunin, V.V, Skarina, T, Brown, G, Yakunin, A.F, Edwards, A.M, Savchenko, A. | | Deposit date: | 2007-11-30 | | Release date: | 2008-12-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Characterization of the Type II Fructose-1,6-bisphosphatase GlpX from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
1ZS7
 
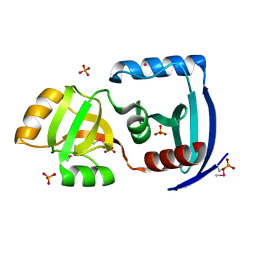 | | The structure of gene product APE0525 from Aeropyrum pernix | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, hypothetical protein APE0525 | | Authors: | Cuff, M.E, Skarina, T, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-05-23 | | Release date: | 2005-07-05 | | Last modified: | 2014-04-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of gene product APE0525 from Aeropyrum pernix
To be Published
|
|
3BIH
 
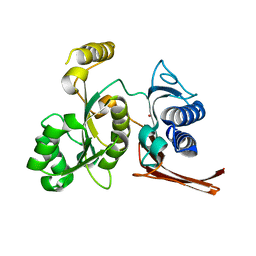 | | Crystal structure of fructose-1,6-bisphosphatase from E.coli GlpX | | Descriptor: | Fructose-1,6-bisphosphatase class II glpX, UNKNOWN ATOM OR ION | | Authors: | Lunin, V.V, Skarina, T, Brown, G, Yakunin, A.F, Edwards, A.M, Savchenko, A. | | Deposit date: | 2007-11-30 | | Release date: | 2008-12-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Biochemical Characterization of the Type II Fructose-1,6-bisphosphatase GlpX from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
3C3J
 
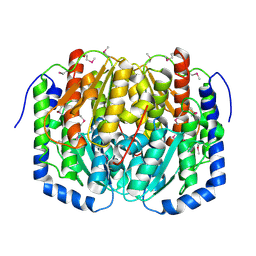 | | Crystal structure of tagatose-6-phosphate ketose/aldose isomerase from Escherichia coli | | Descriptor: | Putative tagatose-6-phosphate ketose/aldose isomerase | | Authors: | Zhang, R, Skarina, T, Egorova, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-28 | | Release date: | 2008-02-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the tagatose-6-phosphate ketose/aldose isomerase from Escherichia coli.
To be Published
|
|
3BQY
 
 | | Crystal structure of a possible TetR family transcriptional regulator from Streptomyces coelicolor A3(2). | | Descriptor: | ACETIC ACID, PHOSPHATE ION, Putative TetR family transcriptional regulator | | Authors: | Cuff, M.E, Skarina, T, Kagan, O, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-20 | | Release date: | 2008-01-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of a possible TetR family transcriptional regulator from Streptomyces coelicolor A3(2).
TO BE PUBLISHED
|
|
