3WPB
 
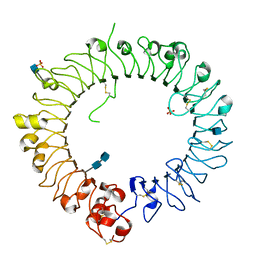 | | Crystal structure of horse TLR9 (unliganded form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | Authors: | Ohto, U, Tanji, H, Shimizu, T. | | Deposit date: | 2014-01-11 | | Release date: | 2015-02-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9
Nature, 520, 2015
|
|
3WF1
 
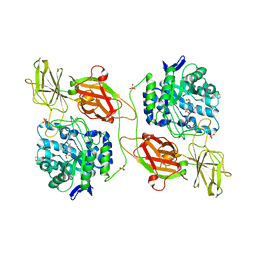 | | Crystal structure of human beta-galactosidase in complex with 6S-NBI-GJ | | Descriptor: | (3E,5S,6R,7S,8S,8aS)-3-(butylimino)hexahydro[1,3]thiazolo[3,4-a]pyridine-5,6,7,8-tetrol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Suzuki, H, Ohto, U, Shimizu, T. | | Deposit date: | 2013-07-16 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of pharmacological chaperoning for human beta-galactosidase
to be published
|
|
3WPD
 
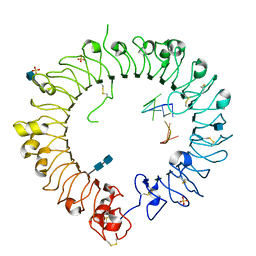 | | Crystal structure of horse TLR9 in complex with inhibitory DNA4084 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-D(*CP*CP*TP*GP*GP*AP*TP*GP*GP*G)-3'), ... | | Authors: | Ohto, U, Tanji, H, Shimizu, T. | | Deposit date: | 2014-01-11 | | Release date: | 2015-02-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9
Nature, 520, 2015
|
|
3W3J
 
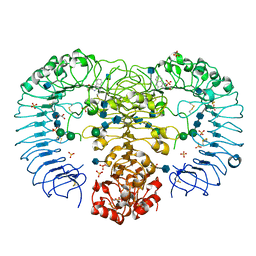 | | Crystal structure of human TLR8 in complex with CL097 | | Descriptor: | 2-(ethoxymethyl)-1H-imidazo[4,5-c]quinolin-4-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2012-12-22 | | Release date: | 2013-04-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural reorganization of the Toll-like receptor 8 dimer induced by agonistic ligands
Science, 339, 2013
|
|
3WPG
 
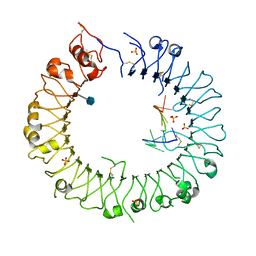 | |
3WF4
 
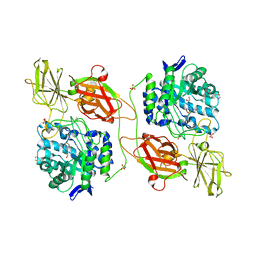 | | Crystal structure of human beta-galactosidase mutant I51T in complex with 6S-NBI-DGJ | | Descriptor: | (3Z,6S,7R,8S,8aS)-3-(butylimino)hexahydro[1,3]thiazolo[3,4-a]pyridine-6,7,8-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Suzuki, H, Ohto, U, Shimizu, T. | | Deposit date: | 2013-07-16 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of pharmacological chaperoning for human beta-galactosidase
to be published
|
|
3WPI
 
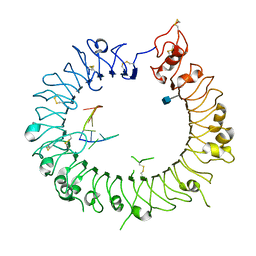 | | Crystal structure of mouse TLR9 in complex with inhibitory DNA_super | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-D(*CP*CP*TP*CP*AP*AP*TP*AP*GP*GP*GP*TP*GP*AP*GP*GP*GP*G)-3'), Toll-like receptor 9 | | Authors: | Ohto, U, Shimizu, T. | | Deposit date: | 2014-01-11 | | Release date: | 2015-02-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9
Nature, 520, 2015
|
|
3W3L
 
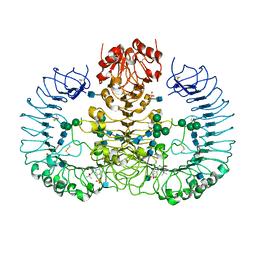 | | Crystal structure of human TLR8 in complex with Resiquimod (R848) crystal form 1 | | Descriptor: | 1-[4-amino-2-(ethoxymethyl)-1H-imidazo[4,5-c]quinolin-1-yl]-2-methylpropan-2-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2012-12-22 | | Release date: | 2013-04-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural reorganization of the Toll-like receptor 8 dimer induced by agonistic ligands
Science, 339, 2013
|
|
3WPE
 
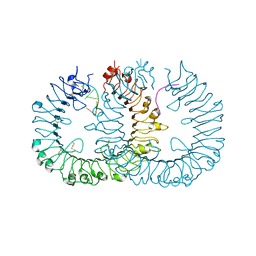 | |
3WF2
 
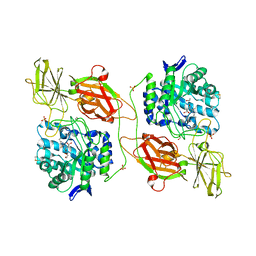 | | Crystal structure of human beta-galactosidase in complex with NBT-DGJ | | Descriptor: | (2R,3S,4R,5S)-N-butyl-3,4,5-trihydroxy-2-(hydroxymethyl)piperidine-1-carbothioamide, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Suzuki, H, Ohto, U, Shimizu, T. | | Deposit date: | 2013-07-16 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of pharmacological chaperoning for human beta-galactosidase
to be published
|
|
3WPH
 
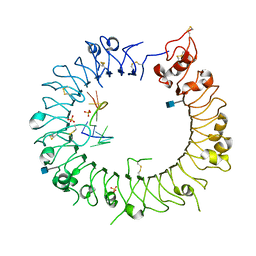 | |
3WF0
 
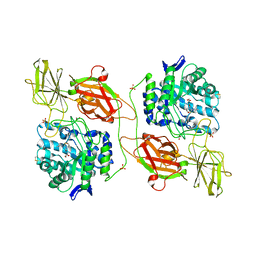 | | Crystal structure of human beta-galactosidase in complex with 6S-NBI-DGJ | | Descriptor: | (3Z,6S,7R,8S,8aS)-3-(butylimino)hexahydro[1,3]thiazolo[3,4-a]pyridine-6,7,8-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Suzuki, H, Ohto, U, Shimizu, T. | | Deposit date: | 2013-07-16 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of pharmacological chaperoning for human beta-galactosidase
to be published
|
|
3WPC
 
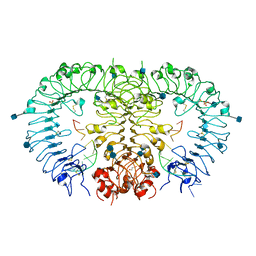 | | Crystal structure of horse TLR9 in complex with agonistic DNA1668_12mer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-D(*CP*AP*TP*GP*AP*CP*GP*TP*TP*CP*CP*T)-3'), ... | | Authors: | Ohto, U, Tanji, H, Shimizu, T. | | Deposit date: | 2014-01-11 | | Release date: | 2015-02-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9
Nature, 520, 2015
|
|
2CT9
 
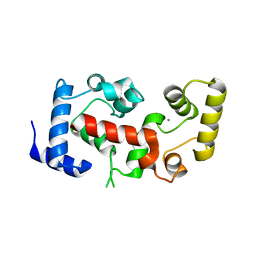 | | The crystal structure of calcineurin B homologous proein 1 (CHP1) | | Descriptor: | CALCIUM ION, Calcium-binding protein p22 | | Authors: | Naoe, Y, Arita, K, Hashimoto, H, Kanazawa, H, Sato, M, Shimizu, T. | | Deposit date: | 2005-05-23 | | Release date: | 2005-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of calcineurin B homologous protein 1
J.Biol.Chem., 280, 2005
|
|
2DEX
 
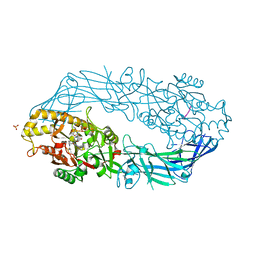 | | Crystal structure of human peptidylarginine deiminase 4 in complex with histone H3 N-terminal peptide including Arg17 | | Descriptor: | 10-mer peptide from histone H3, CALCIUM ION, Protein-arginine deiminase type IV, ... | | Authors: | Arita, K, Shimizu, T, Hashimoto, H, Hidaka, Y, Yamada, M, Sato, M. | | Deposit date: | 2006-02-18 | | Release date: | 2006-04-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for histone N-terminal recognition by human peptidylarginine deiminase 4
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2DEW
 
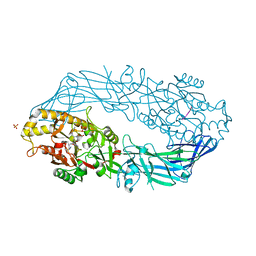 | | Crystal structure of human peptidylarginine deiminase 4 in complex with histone H3 N-terminal tail including Arg8 | | Descriptor: | 10-mer peptide from histone H3, CALCIUM ION, Protein-arginine deiminase type IV, ... | | Authors: | Arita, K, Shimizu, T, Hashimoto, H, Hidaka, Y, Yamada, M, Sato, M. | | Deposit date: | 2006-02-18 | | Release date: | 2006-04-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for histone N-terminal recognition by human peptidylarginine deiminase 4
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2DEY
 
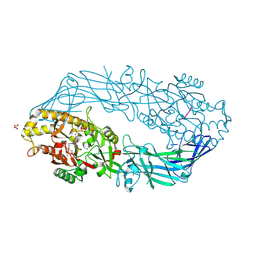 | | Crystal structure of human peptidylarginine deiminase 4 in complex with histone H4 N-terminal tail including Arg3 | | Descriptor: | 10-mer peptide from histone H4, CALCIUM ION, Protein-arginine deiminase type IV, ... | | Authors: | Arita, K, Shimizu, T, Hashimoto, H, Hidaka, Y, Yamada, M, Sato, M. | | Deposit date: | 2006-02-18 | | Release date: | 2006-04-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for histone N-terminal recognition by human peptidylarginine deiminase 4
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2E1N
 
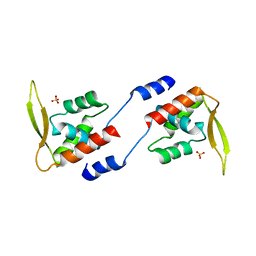 | |
3T6Q
 
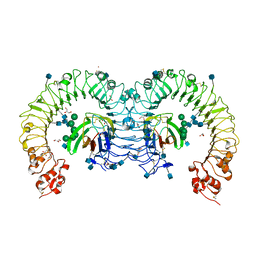 | | Crystal structure of mouse RP105/MD-1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD180 antigen, ... | | Authors: | Ohto, U, Shimizu, T. | | Deposit date: | 2011-07-28 | | Release date: | 2011-11-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Mouse and Human RP105/MD-1 Complexes Reveal Unique Dimer Organization of the Toll-Like Receptor Family.
J.Mol.Biol., 413, 2011
|
|
3VQ1
 
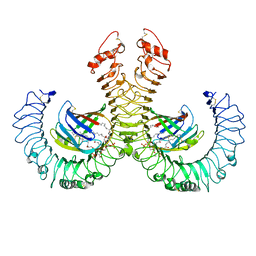 | | Crystal structure of mouse TLR4/MD-2/lipid IVa complex | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ohto, U, Shimizu, T. | | Deposit date: | 2012-03-17 | | Release date: | 2012-05-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of species-specific endotoxin sensing by innate immune receptor TLR4/MD-2
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VU7
 
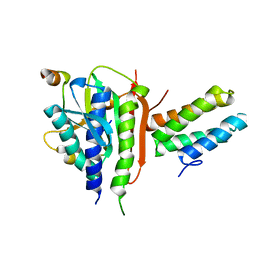 | | Crystal structure of REV1-REV7-REV3 ternary complex | | Descriptor: | DNA polymerase zeta catalytic subunit, DNA repair protein REV1, Mitotic spindle assembly checkpoint protein MAD2B | | Authors: | Kikuchi, S, Hara, K, Shimizu, T, Sato, M, Hashimoto, H. | | Deposit date: | 2012-06-20 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of recruitment of DNA polymerase [zeta] by interaction between REV1 and REV7 proteins
J.Biol.Chem., 287, 2012
|
|
3VQ2
 
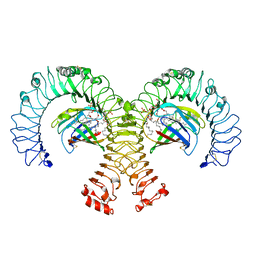 | | Crystal structure of mouse TLR4/MD-2/LPS complex | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ohto, U, Shimizu, T. | | Deposit date: | 2012-03-17 | | Release date: | 2012-05-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural basis of species-specific endotoxin sensing by innate immune receptor TLR4/MD-2
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VIR
 
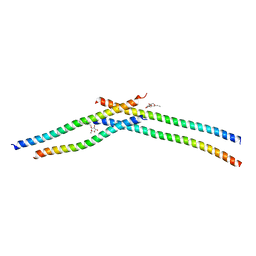 | | Crystal strcture of Swi5 from fission yeast | | Descriptor: | Mating-type switching protein swi5, octyl beta-D-glucopyranoside | | Authors: | Kuwabara, N, Yamada, N, Hashimoto, H, Sato, M, Iwasaki, H, Shimizu, T. | | Deposit date: | 2011-10-06 | | Release date: | 2012-08-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanistic insights into the activation of Rad51-mediated strand exchange from the structure of a recombination activator, the Swi5-Sfr1 complex
Structure, 20, 2012
|
|
3VIQ
 
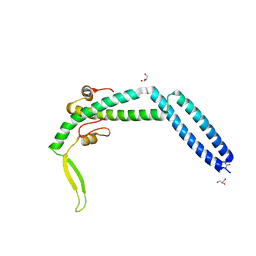 | | Crystal structure of Swi5-Sfr1 complex from fission yeast | | Descriptor: | GLYCEROL, Mating-type switching protein swi5, NITRATE ION, ... | | Authors: | Kuwabara, N, Murayama, Y, Hashimoto, H, Kokabu, Y, Ikeguchi, M, Sato, M, Mayanagi, K, Tsutsui, Y, Iwasaki, H, Shimizu, T. | | Deposit date: | 2011-10-06 | | Release date: | 2012-08-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic insights into the activation of Rad51-mediated strand exchange from the structure of a recombination activator, the Swi5-Sfr1 complex
Structure, 20, 2012
|
|
3WN4
 
 | | Crystal structure of human TLR8 in complex with DS-877 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-butylfuro[2,3-c]quinolin-4-amine, Toll-like receptor 8, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2013-12-02 | | Release date: | 2014-02-19 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure-based design of novel human Toll-like receptor 8 agonists.
Chemmedchem, 9, 2014
|
|
