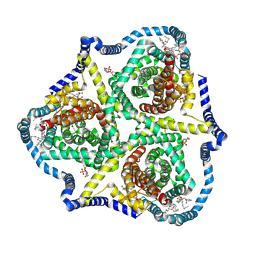
8TZ3
 
 | |
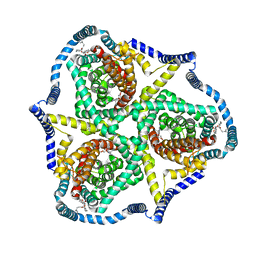
8TZ7
 
 | |
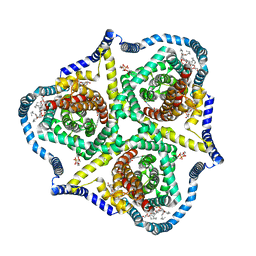
8TZ5
 
 | |
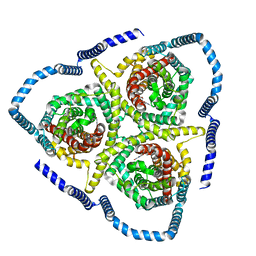
8TZA
 
 | |
8TZ8
 
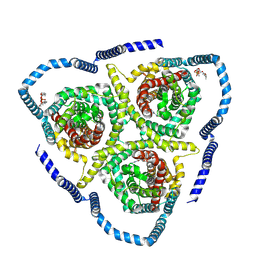 | |
8TZ1
 
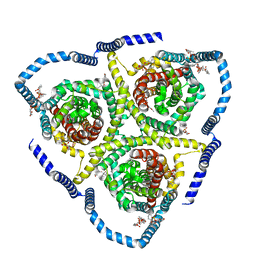 | |
8TZ4
 
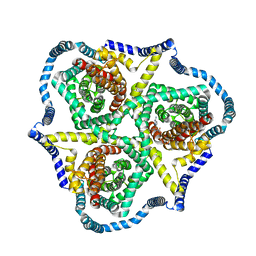 | | Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with GS-441524, subset reconstruction | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(4-azanylpyrrolo[2,1-f][1,2,4]triazin-7-yl)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolane-2-carbonitrile, Sodium/nucleoside cotransporter | | Authors: | Wright, N.J, Lee, S.-Y. | | Deposit date: | 2023-08-26 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Antiviral drug recognition and elevator-type transport motions of CNT3.
Nat.Chem.Biol., 2024
|
|
6FOP
 
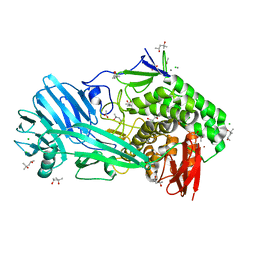 | | Glycoside hydrolase family 81 from Clostridium thermocellum (CtLam81A), Mutant E515A | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Correia, M.A.S.C, Carvalho, A.L. | | Deposit date: | 2018-02-08 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Novel insights into the degradation of beta-1,3-glucans by the cellulosome of Clostridium thermocellum revealed by structure and function studies of a family 81 glycoside hydrolase.
Int.J.Biol.Macromol., 117, 2018
|
|
6Y4Q
 
 | | Structure of a stapled peptide bound to MDM2 | | Descriptor: | ACE-LEU-THR-PHE-GLY-GLU-TYR-TRP-ALA-GLN-LEU-ALA-SER, E3 ubiquitin-protein ligase Mdm2, ~{N}-[(1-ethyl-1,2,3-triazol-4-yl)methyl]-~{N},5-dimethyl-4-[2-[2-methyl-5-[methyl-[(1-propyl-1,2,3-triazol-4-yl)methyl]carbamoyl]thiophen-3-yl]cyclopenten-1-yl]thiophene-2-carboxamide | | Authors: | Pantelejevs, T, Bakanovych, I. | | Deposit date: | 2020-02-22 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Diarylethene moiety as an enthalpy-entropy switch: photoisomerizable stapled peptides for modulating p53/MDM2 interaction.
Org.Biomol.Chem., 18, 2020
|
|
4TXD
 
 | | Crystal structure of Thermofilum pendens Csc2 | | Descriptor: | Csc2, ZINC ION | | Authors: | Hrle, A, Conti, E. | | Deposit date: | 2014-07-03 | | Release date: | 2014-08-27 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.818 Å) | | Cite: | Structural analyses of the CRISPR protein Csc2 reveal the RNA-binding interface of the type I-D Cas7 family.
Rna Biol., 11, 2014
|
|
4Z7L
 
 | | Crystal structure of Cas6b | | Descriptor: | Cas6b, RNA (5'-R(*GP*CP*AP*AP*AP*AP*UP*AP*AP*CP*AP*AP*GP*C)-3'), SULFATE ION | | Authors: | Li, H. | | Deposit date: | 2015-04-07 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | A Non-Stem-Loop CRISPR RNA Is Processed by Dual Binding Cas6.
Structure, 24, 2016
|
|
4Z7K
 
 | |
5HY7
 
 | |
5IFE
 
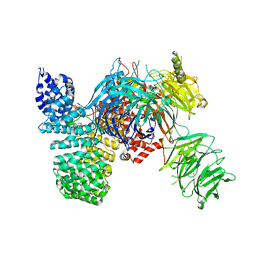 | | Crystal structure of the human SF3b core complex | | Descriptor: | PHD finger-like domain-containing protein 5A, POTASSIUM ION, Splicing factor 3B subunit 1, ... | | Authors: | Cretu, C, Dybkov, O, De Laurentiis, E, Will, C.L, Luhrmann, R, Pena, V. | | Deposit date: | 2016-02-25 | | Release date: | 2016-10-26 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular Architecture of SF3b and Structural Consequences of Its Cancer-Related Mutations.
Mol.Cell, 64, 2016
|
|
4BY6
 
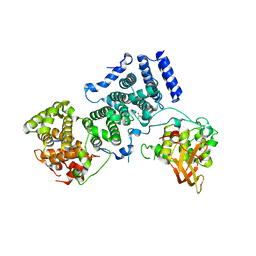 | | Yeast Not1-Not2-Not5 complex | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bhaskar, V, Basquin, J, Conti, E. | | Deposit date: | 2013-07-18 | | Release date: | 2013-10-16 | | Last modified: | 2013-11-20 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Structure and RNA-Binding Properties of the not1-not2-not5 Module of the Yeast Ccr4-not Complex
Nat.Struct.Mol.Biol., 20, 2013
|
|
7F71
 
 | | Crystal structure of the Mycobacterium tuberculosis L,D-transpeptidase-2 (LdtMt2) with peptidoglycan sugar moiety and glutamate | | Descriptor: | GLUTAMIC ACID, GLYCEROL, L,D-transpeptidase 2, ... | | Authors: | Kumar, P, Lamichhane, G. | | Deposit date: | 2021-06-26 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Allosteric cooperation in beta-lactam binding to a non-classical transpeptidase.
Elife, 11, 2022
|
|
7F8P
 
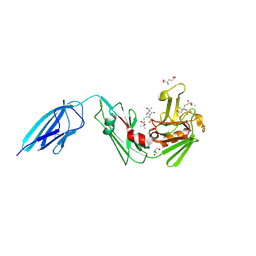 | | Crystal structure of the Mycobacterium tuberculosis L,D-transpeptidase-2 (LdtMt2) with new carbapenem drug T203 | | Descriptor: | (2R,3R)-3-methyl-4-(2-oxidanylidene-2-propan-2-yloxy-ethyl)sulfanyl-2-[(2R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]-2,3-dihydro-1H-pyrrole-5-carboxylic acid, (4R,5S,6S)-6-((R)-1-hydroxyethyl)-3-((2-isopropoxy-2-oxoethyl)thio)-4-methyl-7-oxo-1-azabicyclo[3.2.0]hept-2-ene-2-carboxylic acid, GLUTAMIC ACID, ... | | Authors: | Kumar, P, Lamichhane, G. | | Deposit date: | 2021-07-02 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Allosteric cooperation in beta-lactam binding to a non-classical transpeptidase.
Elife, 11, 2022
|
|
4PCJ
 
 | |
