148D
 
 | |
1K4X
 
 | | POTASSIUM FORM OF OXY-1.5 QUADRUPLEX DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3') | | Authors: | Schultze, P, Hud, N.V, Smith, F.W, Feigon, J. | | Deposit date: | 1999-06-08 | | Release date: | 1999-06-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The effect of sodium, potassium and ammonium ions on the conformation of the dimeric quadruplex formed by the Oxytricha nova telomere repeat oligonucleotide d(G(4)T(4)G(4)).
Nucleic Acids Res., 27, 1999
|
|
156D
 
 | |
7ZVW
 
 | | NuA4 Histone Acetyltransferase Complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Chromatin modification-related protein EAF1, ... | | Authors: | Schultz, P, Ben-Shem, A, Frechard, A. | | Deposit date: | 2022-05-17 | | Release date: | 2023-05-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structure of the NuA4-Tip60 complex reveals the mechanism and importance of long-range chromatin modification.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2MRT
 
 | | CONFORMATION OF CD-7 METALLOTHIONEIN-2 FROM RAT LIVER IN AQUEOUS SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2 | | Authors: | Braun, W, Schultze, P, Woergoetter, E, Wagner, G, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformation of [Cd7]-metallothionein-2 from rat liver in aqueous solution determined by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 203, 1988
|
|
1MRT
 
 | | CONFORMATION OF CD-7 METALLOTHIONEIN-2 FROM RAT LIVER IN AQUEOUS SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2 | | Authors: | Braun, W, Schultze, P, Woergoetter, E, Wagner, G, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformation of [Cd7]-metallothionein-2 from rat liver in aqueous solution determined by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 203, 1988
|
|
1GN7
 
 | |
8OF4
 
 | | Nucleosome Bound human SIRT6 (Composite) | | Descriptor: | DNA (145-MER), Histone H2A type 1, Histone H2B, ... | | Authors: | Smirnova, E, Bignon, E, Schultz, P, Papai, G, Ben-Shem, A. | | Deposit date: | 2023-03-13 | | Release date: | 2023-08-09 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Binding to nucleosome poises human SIRT6 for histone H3 deacetylation.
Elife, 12, 2024
|
|
2QMI
 
 | | Structure of the octameric penicillin-binding protein homologue from Pyrococcus abyssi | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, LUTETIUM (III) ION, Pbp related beta-lactamase | | Authors: | Delfosse, V, Girard, E, Moulinier, L, Schultz, P, Mayer, C. | | Deposit date: | 2007-07-16 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the archaeal pab87 peptidase reveals a novel self-compartmentalizing protease family
Plos One, 4, 2009
|
|
8QR1
 
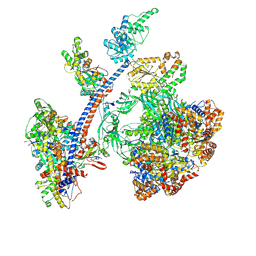 | | Cryo-EM structure of the human Tip60 complex | | Descriptor: | Actin, cytoplasmic 1, N-terminally processed, ... | | Authors: | Li, C, Smirnova, E, Schnitzler, C, Crucifix, C, Concordet, J.P, Brion, A, Poterszman, A, Schultz, P, Papai, G, Ben-Shem, A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-08-07 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure of the human TIP60-C histone exchange and acetyltransferase complex.
Nature, 635, 2024
|
|
8QRI
 
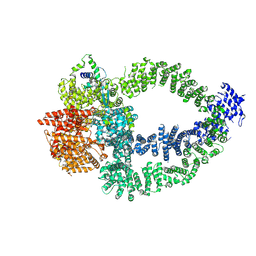 | | TRRAP and EP400 in the human Tip60 complex | | Descriptor: | E1A-binding protein p400, Transformation/transcription domain-associated protein | | Authors: | Li, C, Smirnova, E, Schnitzler, C, Crucifix, C, Concordet, J.P, Brion, A, Poterszman, A, SChultz, P, Papai, G, Ben-Shem, A. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-07 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the human TIP60-C histone exchange and acetyltransferase complex.
Nature, 635, 2024
|
|
5OEJ
 
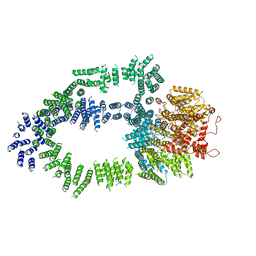 | | Structure of Tra1 subunit within the chromatin modifying complex SAGA | | Descriptor: | Tra1 subunit within the chromatin modifying complex SAGA | | Authors: | Sharov, G, Voltz, K, Durand, A, Kolesnikova, O, Papai, G, Myasnikov, A.G, Dejaegere, A, Ben-Shem, A, Schultz, P. | | Deposit date: | 2017-07-07 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structure of the transcription activator target Tra1 within the chromatin modifying complex SAGA.
Nat Commun, 8, 2017
|
|
6HQA
 
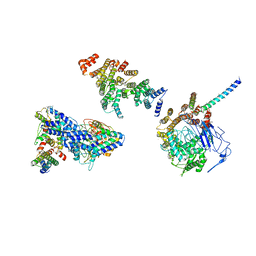 | | Molecular structure of promoter-bound yeast TFIID | | Descriptor: | Histone-fold, Subunit (17 kDa) of TFIID and SAGA complexes, involved in RNA polymerase II transcription initiation, ... | | Authors: | Kolesnikova, O, Ben-Shem, A, Luo, J, Ranish, J, Schultz, P, Papai, G. | | Deposit date: | 2018-09-24 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Molecular structure of promoter-bound yeast TFIID.
Nat Commun, 9, 2018
|
|
6TBM
 
 | | Structure of SAGA bound to TBP, including Spt8 and DUB | | Descriptor: | Polyubiquitin-B, SAGA-associated factor 11, Spt20, ... | | Authors: | Papai, G, Frechard, A, Kolesnikova, O, Crucifix, C, Schultz, P, Ben-Shem, A. | | Deposit date: | 2019-11-01 | | Release date: | 2020-02-12 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Structure of SAGA and mechanism of TBP deposition on gene promoters.
Nature, 577, 2020
|
|
6TB4
 
 | | Structure of SAGA bound to TBP | | Descriptor: | SAGA-associated factor 73 (Sgf73), Spt20, Subunit (17 kDa) of TFIID and SAGA complexes, ... | | Authors: | Papai, G, Frechard, A, Kolesnikova, O, Crucifix, C, Schultz, P, Ben-Shem, A. | | Deposit date: | 2019-10-31 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of SAGA and mechanism of TBP deposition on gene promoters.
Nature, 577, 2020
|
|
1D3X
 
 | | INTRAMOLECULAR DNA TRIPLEX, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*A)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*CP*TP*T)-3'), DNA (5'-D(*TP*TP*CP*TP*CP*TP*CP*T)-3') | | Authors: | Tarkoy, M, Phipps, A.K, Schultze, P, Feigon, J. | | Deposit date: | 1998-02-05 | | Release date: | 1998-05-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an intramolecular DNA triplex linked by hexakis(ethylene glycol) units: d(AGAGAGAA-(EG)6-TTCTCTCT-(EG)6-TCTCTCTT).
Biochemistry, 37, 1998
|
|
230D
 
 | |
1MRB
 
 | | THREE-DIMENSIONAL STRUCTURE OF RABBIT LIVER CD7 METALLOTHIONEIN-2A IN AQUEOUS SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2A | | Authors: | Braun, W, Arseniev, A, Schultze, P, Woergoetter, E, Wagner, G, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of rabbit liver [Cd7]metallothionein-2a in aqueous solution determined by nuclear magnetic resonance.
J.Mol.Biol., 201, 1988
|
|
2MRB
 
 | | THREE-DIMENSIONAL STRUCTURE OF RABBIT LIVER CD-7 METALLOTHIONEIN-2A IN AQUEOUS SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2A | | Authors: | Braun, W, Arseniev, A, Schultze, P, Woergoetter, E, Wagner, G, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of rabbit liver [Cd7]metallothionein-2a in aqueous solution determined by nuclear magnetic resonance.
J.Mol.Biol., 201, 1988
|
|
1P3X
 
 | | INTRAMOLECULAR DNA TRIPLEX WITH 1-PROPYNYL DEOXYURIDINE IN THE THIRD STRAND, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*(PDU)P*CP*(PDU)P*(DCM)P*(PDU)P*CP*(PDU)P*(PDU))-3'), DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*CP*TP*CP*TP*CP*T)-3') | | Authors: | Phipps, A.K, Tarkoy, M, Schultze, P, Feigon, J. | | Deposit date: | 1998-02-05 | | Release date: | 1998-05-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an intramolecular DNA triplex containing 5-(1-propynyl)-2'-deoxyuridine residues in the third strand.
Biochemistry, 37, 1998
|
|
1CG7
 
 | | HMG PROTEIN NHP6A FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | PROTEIN (NON HISTONE PROTEIN 6 A) | | Authors: | Allain, F.H.T, Yen, Y.M, Masse, J.E, Schultze, P, Dieckmann, T, Johnson, R.C, Feigon, J. | | Deposit date: | 1999-03-27 | | Release date: | 1999-10-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HMG protein NHP6A and its interaction with DNA reveals the structural determinants for non-sequence-specific binding.
EMBO J., 18, 1999
|
|
1R3X
 
 | | INTRAMOLECULAR DNA TRIPLEX WITH RNA THIRD STRAND, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*CP*TP*CP*TP*CP*T)-3'), RNA (5'-R(*UP*CP*UP*CP*UP*CP*UP*U)-3') | | Authors: | Gotfredsen, C.H, Schultze, P, Feigon, J. | | Deposit date: | 1998-02-06 | | Release date: | 1998-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of an Intramolecular Pyrimidine-Purine-Pyrimidine Triplex Containing an RNA Third Strand
J.Am.Chem.Soc., 120, 1998
|
|
1JB7
 
 | |
5NL0
 
 | | Crystal structure of a 197-bp palindromic 601L nucleosome in complex with linker histone H1 | | Descriptor: | DNA (197-MER), Histone H1.0-B, Histone H2A type 1, ... | | Authors: | Garcia-Saez, I, Petosa, C, Dimitrov, S. | | Deposit date: | 2017-04-03 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (5.4 Å) | | Cite: | Structure and Dynamics of a 197 bp Nucleosome in Complex with Linker Histone H1.
Mol. Cell, 66, 2017
|
|
5MG3
 
 | | EM fitted model of bacterial holo-translocon | | Descriptor: | Membrane protein insertase YidC, Protein translocase subunit SecD, Protein translocase subunit SecE, ... | | Authors: | Schaffitzel, C, Botte, M. | | Deposit date: | 2016-11-20 | | Release date: | 2016-12-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | A central cavity within the holo-translocon suggests a mechanism for membrane protein insertion.
Sci Rep, 6, 2016
|
|
