2VSX
 
 | | Crystal Structure of a Translation Initiation Complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-DEPENDENT RNA HELICASE EIF4A, EUKARYOTIC INITIATION FACTOR 4F SUBUNIT P150 | | Authors: | Schutz, P, Bumann, M, Oberholzer, A.E, Bieniossek, C, Altmann, M, Trachsel, H, Baumann, U. | | Deposit date: | 2008-04-30 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Yeast Eif4A-Eif4G Complex: An RNA-Helicase Controlled by Protein-Protein Interactions.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3LY5
 
 | | DDX18 dead-domain | | Descriptor: | ATP-dependent RNA helicase DDX18, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Schutz, P, Karlberg, T, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kraulis, P, Kotenyova, T, Kotzsch, A, Markova, N, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-02-26 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Comparative Structural Analysis of Human DEAD-Box RNA Helicases.
Plos One, 5, 2010
|
|
3LLM
 
 | | Crystal Structure Analysis of a RNA Helicase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase A, CACODYLATE ION, ... | | Authors: | Schutz, P, Karlberg, T, Collins, R, Arrowsmith, C.H, Berglund, H, Bountra, C, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kraulis, P, Kotenyova, T, Kotzsch, A, Markova, N, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-29 | | Release date: | 2010-05-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human RNA helicase A (DHX9): structural basis for unselective nucleotide base binding in a DEAD-box variant protein.
J.Mol.Biol., 400, 2010
|
|
3IUY
 
 | | Crystal structure of DDX53 DEAD-box domain | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Probable ATP-dependent RNA helicase DDX53 | | Authors: | Schutz, P, Karlberg, T, Collins, R, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kraulis, P, Kotenyova, T, Kotzsch, A, Markova, N, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-31 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Comparative Structural Analysis of Human DEAD-Box RNA Helicases.
Plos One, 5, 2010
|
|
3P0C
 
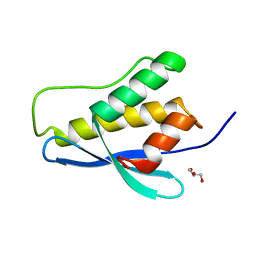 | | Nischarin PX-domain | | Descriptor: | GLYCEROL, Nischarin | | Authors: | Schutz, P, Karlberg, T, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kol, S, Kotenyova, T, Kouznetsova, E, Moche, M, Nyman, T, Persson, C, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Der Berg, S, Wahlberg, E, Welin, M, Weigelt, J, Nordlund, P, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-28 | | Release date: | 2010-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.273 Å) | | Cite: | Crystal structure of Nischarin PX-domain
to be published
|
|
148D
 
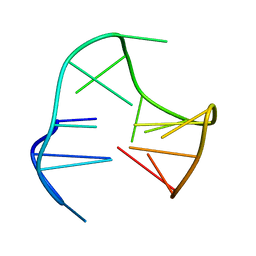 | |
1K4X
 
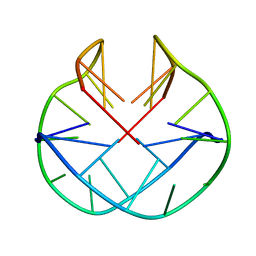 | | POTASSIUM FORM OF OXY-1.5 QUADRUPLEX DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3') | | Authors: | Schultze, P, Hud, N.V, Smith, F.W, Feigon, J. | | Deposit date: | 1999-06-08 | | Release date: | 1999-06-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The effect of sodium, potassium and ammonium ions on the conformation of the dimeric quadruplex formed by the Oxytricha nova telomere repeat oligonucleotide d(G(4)T(4)G(4)).
Nucleic Acids Res., 27, 1999
|
|
2VSO
 
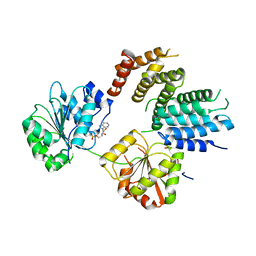 | | Crystal Structure of a Translation Initiation Complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-DEPENDENT RNA HELICASE EIF4A, EUKARYOTIC INITIATION FACTOR 4F SUBUNIT P150 | | Authors: | Schutz, P, Bumann, M, Oberholzer, A.E, Bieniossek, C, Altmann, M, Trachsel, H, Baumann, U. | | Deposit date: | 2008-04-28 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Yeast Eif4A-Eif4G Complex: An RNA-Helicase Controlled by Protein-Protein Interactions.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
156D
 
 | |
7ZVW
 
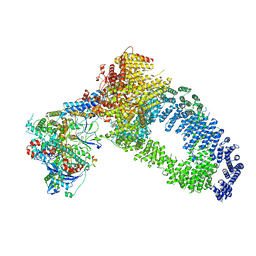 | | NuA4 Histone Acetyltransferase Complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Chromatin modification-related protein EAF1, ... | | Authors: | Schultz, P, Ben-Shem, A, Frechard, A. | | Deposit date: | 2022-05-17 | | Release date: | 2023-05-31 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structure of the NuA4-Tip60 complex reveals the mechanism and importance of long-range chromatin modification.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1MRT
 
 | | CONFORMATION OF CD-7 METALLOTHIONEIN-2 FROM RAT LIVER IN AQUEOUS SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2 | | Authors: | Braun, W, Schultze, P, Woergoetter, E, Wagner, G, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformation of [Cd7]-metallothionein-2 from rat liver in aqueous solution determined by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 203, 1988
|
|
2MRT
 
 | | CONFORMATION OF CD-7 METALLOTHIONEIN-2 FROM RAT LIVER IN AQUEOUS SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2 | | Authors: | Braun, W, Schultze, P, Woergoetter, E, Wagner, G, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformation of [Cd7]-metallothionein-2 from rat liver in aqueous solution determined by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 203, 1988
|
|
1GN7
 
 | |
1NA8
 
 | | Crystal structure of ADP-ribosylation factor binding protein GGA1 | | Descriptor: | ADP-ribosylation factor binding protein GGA1 | | Authors: | Lui, W.W, Collins, B.M, Hirst, J, Motley, A, Millar, C, Schu, P, Owen, D.J, Robinson, M.S. | | Deposit date: | 2002-11-27 | | Release date: | 2003-07-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binding partners for the COOH-terminal appendage domains of the GGAs and gamma-adaptin
Mol.Cell.Biol., 14, 2003
|
|
2IU1
 
 | |
3LS8
 
 | | Crystal structure of human PIK3C3 in complex with 3-[4-(4-Morpholinyl)thieno[3,2-d]pyrimidin-2-yl]-phenol | | Descriptor: | 3-(4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)phenol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tresaugues, L, Welin, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kotenyova, T, Kraulis, P, Moche, M, Nyman, T, Persson, C, Schuler, H, Schutz, P, Siponen, M.I, Thorsell, A.G, Van den Berg, S, Wahlberg, E, Weigelt, J, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-02-12 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of human PIK3C3 in complex with 3-[4-(4-Morpholinyl)thieno[3,2-d]pyrimidin-2-yl]-phenol
To be Published
|
|
3K2I
 
 | | Human Acyl-coenzyme A thioesterase 4 | | Descriptor: | Acyl-coenzyme A thioesterase 4, CHLORIDE ION, NICKEL (II) ION | | Authors: | Siponen, M.I, Moche, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kraulis, P, Kotenyova, T, Kotzsch, A, Markova, N, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Schutz, P, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human Acyl-coenzyme A thioesterase 4
To be Published
|
|
3KVH
 
 | | Crystal structure of human protein syndesmos (NUDT16-like protein) | | Descriptor: | CHLORIDE ION, GLYCEROL, Protein syndesmos | | Authors: | Tresaugues, L, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Kotzsch, A, Kraulis, P, Moche, M, Nielsen, T.K, Nyman, T, Persson, C, Roos, A.K, Schuler, H, Schutz, P, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-30 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human protein syndesmos (NUDT16L1)
To be Published
|
|
3KJD
 
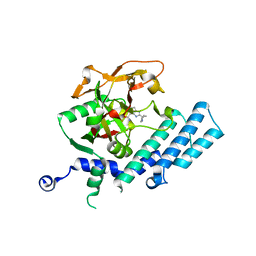 | | Human poly(ADP-ribose) polymerase 2, catalytic fragment in complex with an inhibitor ABT-888 | | Descriptor: | (2R)-2-(7-carbamoyl-1H-benzimidazol-2-yl)-2-methylpyrrolidinium, GLYCEROL, Poly [ADP-ribose] polymerase 2 | | Authors: | Karlberg, T, Schutz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kotenyova, T, Kotzsch, A, Kraulis, P, Nielsen, T.K, Moche, M, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-03 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the catalytic domain of human PARP2 in complex with PARP inhibitor ABT-888.
Biochemistry, 49, 2010
|
|
3KCZ
 
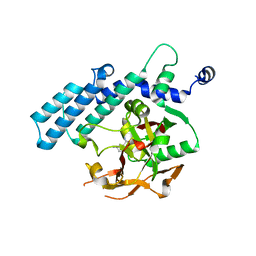 | | Human poly(ADP-ribose) polymerase 2, catalytic fragment in complex with an inhibitor 3-aminobenzamide | | Descriptor: | 3-aminobenzamide, GLYCEROL, Poly [ADP-ribose] polymerase 2 | | Authors: | Karlberg, T, Schutz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kotenyova, T, Kotzsch, A, Kraulis, P, Nielsen, T.K, Moche, M, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-22 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the catalytic domain of human PARP2 in complex with PARP inhibitor ABT-888.
Biochemistry, 49, 2010
|
|
3KR8
 
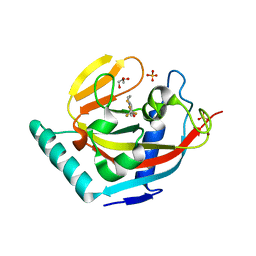 | | Human tankyrase 2 - catalytic PARP domain in complex with an inhibitor XAV939 | | Descriptor: | 2-[4-(trifluoromethyl)phenyl]-7,8-dihydro-5H-thiopyrano[4,3-d]pyrimidin-4-ol, GLYCEROL, SULFATE ION, ... | | Authors: | Karlberg, T, Schutz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kotenyova, T, Kotzsch, A, Kraulis, P, Nielsen, T.K, Moche, M, Nordlund, P, Nyman, T, Persson, C, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-18 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the interaction between tankyrase-2 and a potent Wnt-signaling inhibitor.
J.Med.Chem., 53, 2010
|
|
3KR7
 
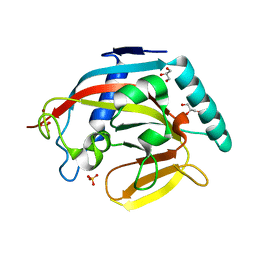 | | Human tankyrase 2 - catalytic PARP domain | | Descriptor: | GLYCEROL, SULFATE ION, Tankyrase-2, ... | | Authors: | Karlberg, T, Schutz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kotenyova, T, Kotzsch, A, Kraulis, P, Nielsen, T.K, Moche, M, Nordlund, P, Nyman, T, Persson, C, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-18 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the interaction between tankyrase-2 and a potent Wnt-signaling inhibitor.
J.Med.Chem., 53, 2010
|
|
3MCF
 
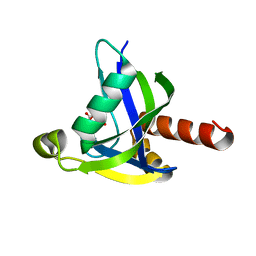 | | Crystal structure of human diphosphoinositol polyphosphate phosphohydrolase 3-alpha | | Descriptor: | CITRATE ANION, Diphosphoinositol polyphosphate phosphohydrolase 3-alpha, GLYCEROL | | Authors: | Tresaugues, L, Welin, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kol, S, Kotenyova, T, Moche, M, Nyman, T, Persson, C, Schuler, H, Schutz, P, Siponen, M.I, Thorsell, A.G, van der Berg, S, Wahlberg, E, Weigelt, J, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-29 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human diphosphoinositol polyphosphate phosphohydrolase 3-alpha
To be Published
|
|
3MTG
 
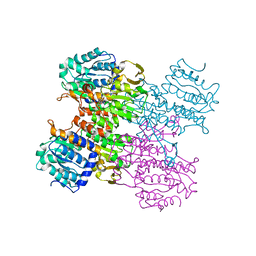 | | Crystal structure of human S-adenosyl homocysteine hydrolase-like 1 protein | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative adenosylhomocysteinase 2 | | Authors: | Wisniewska, M, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Schutz, P, Thorsell, A.G, Tresaugues, L, van der Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-30 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of human S-adenosyl homocysteine hydrolase-like 1 protein
TO BE PUBLISHED
|
|
3N8E
 
 | | Substrate binding domain of the human Heat Shock 70kDa protein 9 (mortalin) | | Descriptor: | Stress-70 protein, mitochondrial | | Authors: | Wisniewska, M, Karlberg, T, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Schutz, P, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, van der Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-28 | | Release date: | 2010-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Substrate binding domain of the human Heat Shock 70kDa protein 9 (mortalin)
To be published
|
|
