1I2R
 
 | |
1DAM
 
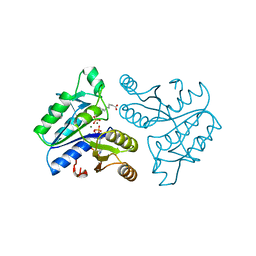 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH DETHIOBIOTIN, ADP, INORGANIC PHOSPHATE AND MAGNESIUM | | Descriptor: | 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kaeck, H, Sandmark, J, Gibson, K.J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1998-08-31 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of two quaternary complexes of dethiobiotin synthetase, enzyme-MgADP-AlF3-diaminopelargonic acid and enzyme-MgADP-dethiobiotin-phosphate; implications for catalysis.
Protein Sci., 7, 1998
|
|
2F74
 
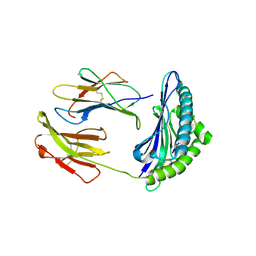 | | Murine MHC class I H-2Db in complex with human b2-microglobulin and LCMV-derived immunodminant peptide gp33 | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Achour, A, Michaelsson, J, Harris, R.A, Ljunggren, H.G, Karre, K, Schneider, G, Sandalova, T. | | Deposit date: | 2005-11-30 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of the Differential Stability and Receptor Specificity of H-2D(b) in Complex with Murine versus Human beta(2)-Microglobulin.
J.Mol.Biol., 356, 2006
|
|
1GPU
 
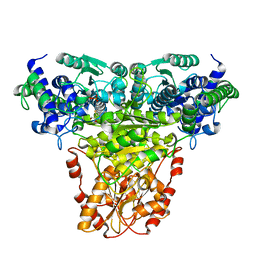 | | Transketolase complex with reaction intermediate | | Descriptor: | 2-[3-[(4-AMINO-2-METHYL-5-PYRIMIDINYL)METHYL]-2-(1,2-DIHYDROXYETHYL)-4-METHYL-1,3-THIAZOL-3-IUM-5-YL]ETHYL TRIHYDROGEN DIPHOSPHATE, CALCIUM ION, TRANSKETOLASE 1 | | Authors: | Fiedler, E, Thorell, S, Sandalova, T, Koenig, S, Schneider, G. | | Deposit date: | 2001-11-09 | | Release date: | 2002-02-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Snapshot of a Key Intermediate in Enzymatic Thiamin Catalysis: Crystal Structure of the Alpha-Carbanion of (Alpha,Beta-Dihydroxyethyl)-Thiamin Diphosphate in the Active Site of Transketolase from Saccharomyces Cerevisiae.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2Y5T
 
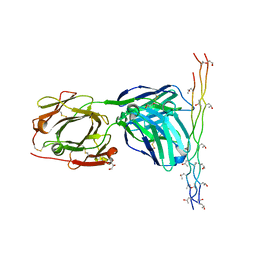 | | Crystal structure of the pathogenic autoantibody CIIC1 in complex with the triple-helical C1 peptide | | Descriptor: | C1, CHLORIDE ION, CIIC1 FAB FRAGMENT HEAVY CHAIN, ... | | Authors: | Dobritzsch, D, Lindh, I, Schneider, N, Uysal, H, Nandakumar, K.S, Burkhardt, H, Schneider, G, Holmdahl, R. | | Deposit date: | 2011-01-17 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of an Arthritogenic Anticollagen Immune Complex.
Arthritis Rheum., 63, 2011
|
|
8PBO
 
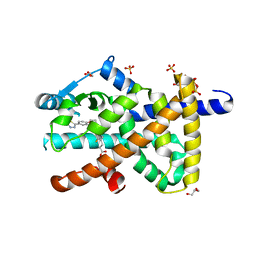 | | Deep interactome learning for generative drug design | | Descriptor: | 3-[2-fluoranyl-4-[3-[2-fluoranyl-4-(5-methyl-1,3,4-thiadiazol-2-yl)phenoxy]propoxy]phenyl]propanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor gamma, ... | | Authors: | Hakansson, M, Focht, D, Atz, K, Schneider, G. | | Deposit date: | 2023-06-09 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Prospective de novo drug design with deep interactome learning.
Nat Commun, 15, 2024
|
|
1NGS
 
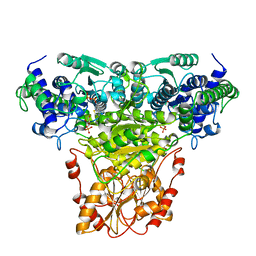 | | COMPLEX OF TRANSKETOLASE WITH THIAMIN DIPHOSPHATE, CA2+ AND ACCEPTOR SUBSTRATE ERYTHROSE-4-PHOSPHATE | | Descriptor: | CALCIUM ION, ERYTHOSE-4-PHOSPHATE, THIAMINE DIPHOSPHATE, ... | | Authors: | Nilsson, U, Lindqvist, Y, Schneider, G. | | Deposit date: | 1996-09-25 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Examination of substrate binding in thiamin diphosphate-dependent transketolase by protein crystallography and site-directed mutagenesis.
J.Biol.Chem., 272, 1997
|
|
1YBV
 
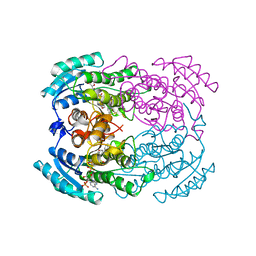 | | STRUCTURE OF TRIHYDROXYNAPHTHALENE REDUCTASE IN COMPLEX WITH NADPH AND AN ACTIVE SITE INHIBITOR | | Descriptor: | 5-METHYL-1,2,4-TRIAZOLO[3,4-B]BENZOTHIAZOLE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRIHYDROXYNAPHTHALENE REDUCTASE | | Authors: | Andersson, A, Schneider, G, Lindqvist, Y. | | Deposit date: | 1996-09-23 | | Release date: | 1997-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the ternary complex of 1,3,8-trihydroxynaphthalene reductase from Magnaporthe grisea with NADPH and an active-site inhibitor.
Structure, 4, 1996
|
|
1ZPD
 
 | | PYRUVATE DECARBOXYLASE FROM ZYMOMONAS MOBILIS | | Descriptor: | CITRIC ACID, MAGNESIUM ION, MONO-{4-[(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-AMINO]-2-HYDROXY-3-MERCAPTO-PENT-3-ENYL-PHOSPHONO} ESTER, ... | | Authors: | Lu, G, Dobritzsch, D, Schneider, G. | | Deposit date: | 1998-04-17 | | Release date: | 1999-02-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | High resolution crystal structure of pyruvate decarboxylase from Zymomonas mobilis. Implications for substrate activation in pyruvate decarboxylases.
J.Biol.Chem., 273, 1998
|
|
1ZHB
 
 | | Crystal Structure Of The Murine Class I Major Histocompatibility Complex Of H-2Db, B2-Microglobulin, and a 9-Residue Peptide Derived from rat dopamine beta-monooxigenase | | Descriptor: | 9-mer peptide from Dopamine beta-monooxygenase, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Sandalova, T, Michaelsson, J, Harris, R.A, Odeberg, J, Schneider, G, Karre, K, Achour, A. | | Deposit date: | 2005-04-25 | | Release date: | 2005-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A structural basis for CD8+ T cell-dependent recognition of non-homologous peptide ligands: implications for molecular mimicry in autoreactivity
J.Biol.Chem., 280, 2005
|
|
2N8D
 
 | |
3CWN
 
 | | Escherichia coli transaldolase b mutant f178y | | Descriptor: | SULFATE ION, Transaldolase B | | Authors: | Sandalova, T, Schneider, G, Samland, A. | | Deposit date: | 2008-04-22 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Replacement of a Phenylalanine by a Tyrosine in the Active Site Confers Fructose-6-phosphate Aldolase Activity to the Transaldolase of Escherichia coli and Human Origin.
J.Biol.Chem., 283, 2008
|
|
3KG1
 
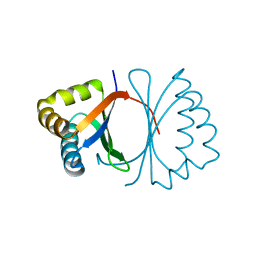 | | Crystal structure of SnoaB, a cofactor-independent oxygenase from Streptomyces nogalater, mutant N63A | | Descriptor: | CHLORIDE ION, SnoaB | | Authors: | Koskiniemi, H, Grocholski, T, Lindqvist, Y, Mantsala, P, Niemi, J, Schneider, G. | | Deposit date: | 2009-10-28 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism
Biochemistry, 49, 2010
|
|
3KG0
 
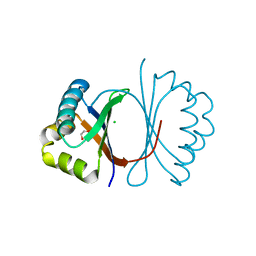 | | Crystal structure of SnoaB, a cofactor-independent oxygenase from Streptomyces nogalater, determined to 1.7 resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SnoaB | | Authors: | Koskiniemi, H, Grocholski, T, Lindqvist, Y, Mantsala, P, Niemi, J, Schneider, G. | | Deposit date: | 2009-10-28 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism
Biochemistry, 49, 2010
|
|
3KNG
 
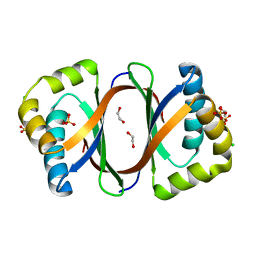 | | Crystal structure of SnoaB, a cofactor-independent oxygenase from Streptomyces nogalater, determined to 1.9 resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Koskiniemi, H, Grocholski, T, Lindqvist, Y, Mantsala, P, Niemi, J, Schneider, G. | | Deposit date: | 2009-11-12 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism
Biochemistry, 49, 2010
|
|
1BYI
 
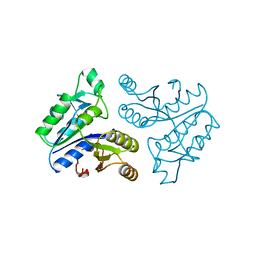 | | STRUCTURE OF APO-DETHIOBIOTIN SYNTHASE AT 0.97 ANGSTROMS RESOLUTION | | Descriptor: | DETHIOBIOTIN SYNTHASE | | Authors: | Sandalova, T, Schneider, G, Kaeck, H, Lindqvist, Y. | | Deposit date: | 1998-10-15 | | Release date: | 1999-06-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Structure of dethiobiotin synthetase at 0.97 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1DAI
 
 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID | | Descriptor: | 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID, DETHIOBIOTIN SYNTHETASE | | Authors: | Huang, W, Jia, J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mechanism of an ATP-dependent carboxylase, dethiobiotin synthetase, based on crystallographic studies of complexes with substrates and a reaction intermediate.
Biochemistry, 34, 1995
|
|
1AY0
 
 | | IDENTIFICATION OF CATALYTICALLY IMPORTANT RESIDUES IN YEAST TRANSKETOLASE | | Descriptor: | CALCIUM ION, THIAMINE DIPHOSPHATE, TRANSKETOLASE | | Authors: | Wikner, C, Nilsson, U, Meshalkina, L, Udekwu, C, Lindqvist, Y, Schneider, G. | | Deposit date: | 1997-11-13 | | Release date: | 1998-05-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of catalytically important residues in yeast transketolase.
Biochemistry, 36, 1997
|
|
2V3W
 
 | | Crystal structure of the benzoylformate decarboxylase variant L461A from Pseudomonas putida | | Descriptor: | BENZOYLFORMATE DECARBOXYLASE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Gocke, D, Walter, L, Gauchenova, K, Kolter, G, Knoll, M, Berthold, C.L, Schneider, G, Pleiss, J, Mueller, M, Pohl, M. | | Deposit date: | 2007-06-25 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Protein Design of Thdp-Dependent Enzymes-Engineering Stereoselectivity.
Chembiochem, 9, 2008
|
|
1AFR
 
 | |
1BII
 
 | | THE CRYSTAL STRUCTURE OF H-2DD MHC CLASS I IN COMPLEX WITH THE HIV-1 DERIVED PEPTIDE P18-110 | | Descriptor: | BETA-2 MICROGLOBULIN, DECAMERIC PEPTIDE, MHC CLASS I H-2DD | | Authors: | Achour, A, Persson, K, Harris, R.A, Sundback, J, Sentman, C.L, Lindqvist, Y, Schneider, G, Karre, K. | | Deposit date: | 1998-06-11 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of H-2Dd MHC class I complexed with the HIV-1-derived peptide P18-I10 at 2.4 A resolution: implications for T cell and NK cell recognition.
Immunity, 9, 1998
|
|
4S2C
 
 | | Covalent complex of E. coli transaldolase TalB with fructose-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, FRUCTOSE -6-PHOSPHATE, Transaldolase B | | Authors: | Stellmacher, L, Sandalova, T, Schneider, G, Sprenger, G.A, Samland, A.K. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel mode of inhibition by D-tagatose 6-phosphate through a Heyns rearrangement in the active site of transaldolase B variants.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
1TRK
 
 | |
4S2B
 
 | | Covalent complex of E. coli transaldolase TalB with tagatose-6-phosphate | | Descriptor: | 2-deoxy-6-O-phosphono-beta-D-lyxo-hexofuranose, SULFATE ION, Transaldolase B | | Authors: | Stellmacher, L, Sandalova, T, Schneider, G, Sprenger, G.A, Samland, A.K. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Novel mode of inhibition by D-tagatose 6-phosphate through a Heyns rearrangement in the active site of transaldolase B variants.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
3CQ5
 
 | | Histidinol-phosphate aminotransferase from Corynebacterium glutamicum in complex with PMP | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Histidinol-phosphate aminotransferase, SULFATE ION, ... | | Authors: | Sandalova, T, Marienhagen, J, Schneider, G. | | Deposit date: | 2008-04-02 | | Release date: | 2008-07-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the structural basis of substrate recognition by histidinol-phosphate aminotransferase from Corynebacterium glutamicum
Acta Crystallogr.,Sect.D, 64, 2008
|
|
