2X7O
 
 | | Crystal structure of TGFbRI complexed with an indolinone inhibitor | | Descriptor: | (3Z)-N-ETHYL-N-METHYL-2-OXO-3-(PHENYL{[4-(PIPERIDIN-1-YLMETHYL)PHENYL]AMINO}METHYLIDENE)-2,3-DIHYDRO-1H-INDOLE-6-CARBOXAMIDE, TGF-BETA RECEPTOR TYPE I | | Authors: | Roth, G.J, Heckel, A, Brandl, T, Grauert, M, Hoerer, S, Kley, J.T, Schnapp, G, Baum, P, Mennerich, D, Schnapp, A, Park, J.E. | | Deposit date: | 2010-03-03 | | Release date: | 2010-10-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Design, Synthesis and Evaluation of Indolinones as Inhibitors of the Transforming Growth Factor Beta Receptor I (Tgfbri)
J.Med.Chem., 53, 2010
|
|
9EOT
 
 | | Structure of human ceramide synthase 6 (CerS6) bound to C16:0 (nanobody Nb02) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 2 of Ceramide synthase 6, ... | | Authors: | Pascoa, T.C, Pike, A.C.W, Chi, G, Stefanic, S, Quigley, A, Chalk, R, Mukhopadhyay, S.M.M, Venkaya, S, Dix, C, Moreira, T, Tessitore, A, Cole, V, Chu, A, Elkins, J.M, Pautsch, A, Schnapp, G, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2024-03-15 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis of the mechanism and inhibition of a human ceramide synthase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
6Y0F
 
 | | Structure of human FAPalpha in complex with linagliptin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 8-[(3R)-3-Aminopiperidin-1-yl]-7-but-2-yn-1-yl-3-methyl-1-[(4-methylquinazolin-2-yl)methyl]-3,7-dihydro-1H-purine-2,6-d ione, ... | | Authors: | Nar, H, Schnapp, G, Schreiner, P. | | Deposit date: | 2020-02-07 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.924 Å) | | Cite: | Structure of human FAPalpha in complex with linagliptin
To Be Published
|
|
8QZ6
 
 | | Structure of human ceramide synthase 6 (CerS6) bound to C16:0 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 2 of Ceramide synthase 6, ... | | Authors: | Pascoa, T.C, Pike, A.C.W, Chi, G, Stefanic, S, Quigley, A, Chalk, R, Mukhopadhyay, S.M.M, Venkaya, S, Dix, C, Moreira, T, Tessitore, A, Cole, V, Chu, A, Elkins, J.M, Pautsch, A, Schnapp, G, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-10-26 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of the mechanism and inhibition of a human ceramide synthase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8QZ7
 
 | | Structure of human ceramide synthase 6 (CerS6) in complex with N-palmitoyl fumonisin B1 | | Descriptor: | (2~{R})-2-[2-[(5~{R},6~{R},7~{S},9~{S},11~{R},16~{R},18~{S},19~{S})-6-[(3~{R})-3-carboxy-5-oxidanyl-5-oxidanylidene-pentanoyl]oxy-19-(hexadecanoylamino)-5,9-dimethyl-11,16,18-tris(oxidanyl)icosan-7-yl]oxy-2-oxidanylidene-ethyl]butanedioic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pascoa, T.C, Pike, A.C.W, Chi, G, Stefanic, S, Quigley, A, Chalk, R, Mukhopadhyay, S.M.M, Venkaya, S, Dix, C, Moreira, T, Tessitore, A, Cole, V, Chu, A, Elkins, J.M, Pautsch, A, Schnapp, G, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-10-26 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of the mechanism and inhibition of a human ceramide synthase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
6TL9
 
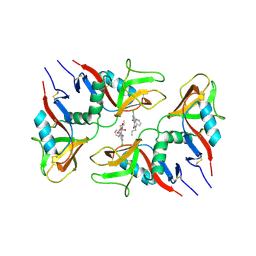 | | CRYSTAL STRUCTURE OF LECTIN-LIKE OX-LDL RECEPTOR 1 IN COMPLEX WITH BI-0115 | | Descriptor: | 9-chloranyl-5-propyl-11~{H}-pyrido[2,3-b][1,4]benzodiazepin-6-one, GLYCEROL, Oxidized low-density lipoprotein receptor 1 | | Authors: | Nar, H, Fiegen, D, Schnapp, G. | | Deposit date: | 2019-12-02 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.734 Å) | | Cite: | A small-molecule inhibitor of lectin-like oxidized LDL receptor-1 acts by stabilizing an inactive receptor tetramer state
Commun Chem, 2020
|
|
6TLA
 
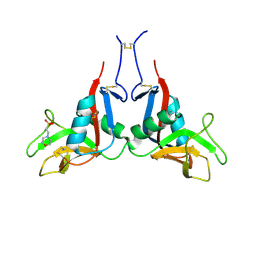 | | CRYSTAL STRUCTURE OF LECTIN-LIKE OX-LDL RECEPTOR 1 (C 1 2 1) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Oxidized low-density lipoprotein receptor 1 | | Authors: | Nar, H, Fiegen, D, Schnapp, G. | | Deposit date: | 2019-12-02 | | Release date: | 2020-07-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A small-molecule inhibitor of lectin-like oxidized LDL receptor-1 acts by stabilizing an inactive receptor tetramer state
Commun Chem, 2020
|
|
6TL7
 
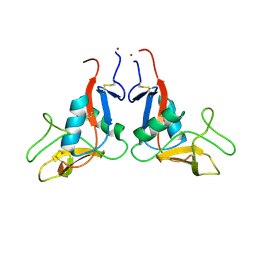 | |
6GPS
 
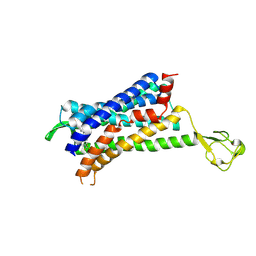 | | CRYSTAL STRUCTURE OF CCR2A IN COMPLEX WITH MK-0812 | | Descriptor: | C-C chemokine receptor type 2,Rubredoxin,C-C chemokine receptor type 2, ZINC ION, [(3~{S},4~{S})-3-methoxyoxan-4-yl]-[(1~{R},3~{S})-3-propan-2-yl-3-[[3-(trifluoromethyl)-7,8-dihydro-5~{H}-1,6-naphthyridin-6-yl]carbonyl]cyclopentyl]azanium | | Authors: | Pautsch, A, Schnapp, G. | | Deposit date: | 2018-06-07 | | Release date: | 2019-01-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of CC Chemokine Receptor 2A in Complex with an Orthosteric Antagonist Provides Insights for the Design of Selective Antagonists.
Structure, 27, 2019
|
|
6GPX
 
 | | CRYSTAL STRUCTURE OF CCR2A IN COMPLEX WITH MK-0812 | | Descriptor: | C-C chemokine receptor type 2,Rubredoxin,C-C chemokine receptor type 2, OLEIC ACID, ZINC ION, ... | | Authors: | Pautsch, A, Schnapp, G, Cheng, R, Apel, A. | | Deposit date: | 2018-06-07 | | Release date: | 2019-01-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of CC Chemokine Receptor 2A in Complex with an Orthosteric Antagonist Provides Insights for the Design of Selective Antagonists.
Structure, 27, 2019
|
|
6H8S
 
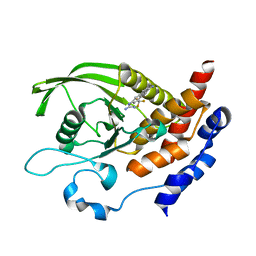 | |
6H8R
 
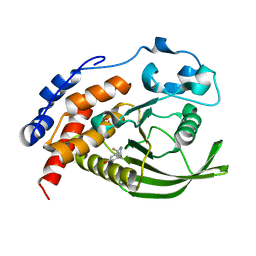 | | CRYSTAL STRUCTURE OF THE HUMAN PROTEIN TYROSINE PHOSPHATASE PTPN5 (STEP) IN COMPLEX WITH COMPOUND 2 | | Descriptor: | 3-[(2~{S})-2-azanylpropyl]-5-(trifluoromethyl)phenol, SULFATE ION, Tyrosine-protein phosphatase non-receptor type 5 | | Authors: | Hoerer, S, Fiegen, D, Schnapp, G. | | Deposit date: | 2018-08-03 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Allosteric Activation of Striatal-Enriched Protein Tyrosine Phosphatase (STEP, PTPN5) by a Fragment-like Molecule.
J. Med. Chem., 62, 2019
|
|
4BBA
 
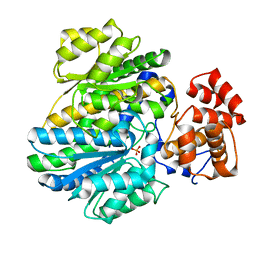 | | Crystal structure of glucokinase regulatory protein complexed to phosphate | | Descriptor: | GLUCOKINASE REGULATORY PROTEIN, PHOSPHATE ION | | Authors: | Pautsch, A, Stadler, N, Loehle, A, Lenter, M, Rist, W, Berg, A, Glocker, L, Nar, H, Reinhart, D, Heckel, A, Schnapp, G, Kauschke, S.G. | | Deposit date: | 2012-09-21 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of Glucokinase Regulatory Protein.
Biochemistry, 52, 2013
|
|
4BB9
 
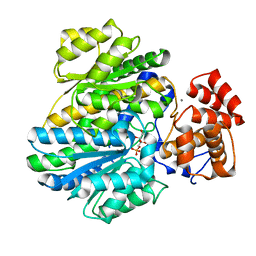 | | Crystal structure of glucokinase regulatory protein complexed to fructose-1-phosphate | | Descriptor: | 1-O-phosphono-beta-D-fructopyranose, CALCIUM ION, GLUCOKINASE REGULATORY PROTEIN | | Authors: | Pautsch, A, Stadler, N, Loehle, A, Lenter, M, Rist, W, Berg, A, Glocker, L, Nar, H, Reinert, D, Heckel, A, Schnapp, G, Kauschke, S.G. | | Deposit date: | 2012-09-21 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal Structure of Glucokinase Regulatory Protein.
Biochemistry, 52, 2013
|
|
5CSM
 
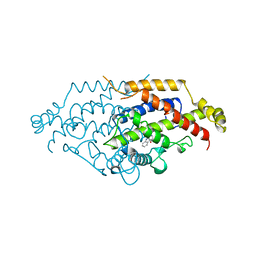 | | YEAST CHORISMATE MUTASE, T226S MUTANT, COMPLEX WITH TRP | | Descriptor: | CHORISMATE MUTASE, TRYPTOPHAN | | Authors: | Straeter, N, Schnappauf, G, Braus, G, Lipscomb, W.N. | | Deposit date: | 1997-07-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanisms of catalysis and allosteric regulation of yeast chorismate mutase from crystal structures.
Structure, 5, 1997
|
|
3CSM
 
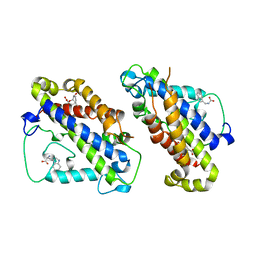 | | STRUCTURE OF YEAST CHORISMATE MUTASE WITH BOUND TRP AND AN ENDOOXABICYCLIC INHIBITOR | | Descriptor: | 8-HYDROXY-2-OXA-BICYCLO[3.3.1]NON-6-ENE-3,5-DICARBOXYLIC ACID, CHORISMATE MUTASE, TRYPTOPHAN | | Authors: | Straeter, N, Schnappauf, G, Braus, G, Lipscomb, W.N. | | Deposit date: | 1997-07-10 | | Release date: | 1998-01-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanisms of catalysis and allosteric regulation of yeast chorismate mutase from crystal structures.
Structure, 5, 1997
|
|
4CSM
 
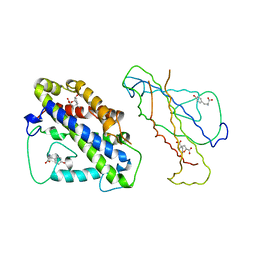 | | YEAST CHORISMATE MUTASE + TYR + ENDOOXABICYCLIC INHIBITOR | | Descriptor: | 8-HYDROXY-2-OXA-BICYCLO[3.3.1]NON-6-ENE-3,5-DICARBOXYLIC ACID, CHORISMATE MUTASE, TYROSINE | | Authors: | Straeter, N, Schnappauf, G, Braus, G, Lipscomb, W.N. | | Deposit date: | 1997-07-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanisms of catalysis and allosteric regulation of yeast chorismate mutase from crystal structures.
Structure, 5, 1997
|
|
5LLS
 
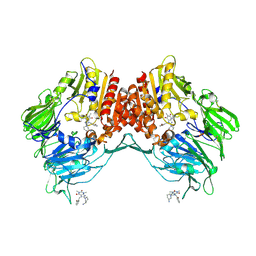 | | Porcine dipeptidyl peptidase IV in complex with 8-(3-aminopiperidin-1-yl)-7-[(2-bromophenyl)methyl]-1,3-dimethyl-2,3,6,7-tetrahydro-1H-purine-2,6-dione | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 8-[(3~{R})-3-azanylpiperidin-1-yl]-7-[(2-bromophenyl)methyl]-1,3-dimethyl-purine-2,6-dione, ... | | Authors: | Nar, H, Blaesse, M. | | Deposit date: | 2016-07-28 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Comparative Analysis of Binding Kinetics and Thermodynamics of Dipeptidyl Peptidase-4 Inhibitors and Their Relationship to Structure.
J.Med.Chem., 59, 2016
|
|
8Q6N
 
 | | Xanthin riboswitch in complex with oxypurinol | | Descriptor: | Oxypurinol, Xanthin riboswitch NMT-46 (46-MER) | | Authors: | Nar, H. | | Deposit date: | 2023-08-14 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Artificial oxypurinol-responsive riboswitches for gene expression control in mammalian cells based on bacterial xanthine aptamers
To Be Published
|
|
8CNI
 
 | | PHT1 in the outward facing conformation, bound to Sb27 | | Descriptor: | Solute carrier family 15 member 4, Sybody 27 | | Authors: | Custodio, T, Killer, M, Loew, C. | | Deposit date: | 2023-02-23 | | Release date: | 2023-09-27 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Molecular basis of TASL recruitment by the peptide/histidine transporter 1, PHT1.
Nat Commun, 14, 2023
|
|
4WBX
 
 | | Conserved hypothetical protein PF1771 from Pyrococcus furiosus solved by sulfur SAD using Swiss Light Source data | | Descriptor: | 2-keto acid:ferredoxin oxidoreductase subunit alpha | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-09-04 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
5HES
 
 | | Human leucine zipper- and sterile alpha motif-containing kinase (ZAK, MLT, HCCS-4, MRK, AZK, MLTK) in complex with vemurafenib | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase kinase kinase MLT, N-(3-{[5-(4-chlorophenyl)-1H-pyrrolo[2,3-b]pyridin-3-yl]carbonyl}-2,4-difluorophenyl)propane-1-sulfonamide | | Authors: | Mathea, S, Salah, E, Abdul Azeez, K.R, Tallant, C, Szklarz, M, Chaikuad, A, Shrestha, B, Sorrell, F.J, Elkins, J.M, Shrestha, L, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2016-01-06 | | Release date: | 2016-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure of the Human Protein Kinase ZAK in Complex with Vemurafenib.
Acs Chem.Biol., 11, 2016
|
|
7XK8
 
 | | Cryo-EM structure of the Neuromedin U receptor 2 (NMUR2) in complex with G Protein and its endogeneous Peptide-Agonist NMU25 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhao, W, Wenru, Z, Mu, W, Minmin, L, Shutian, C, Tingting, T, Gisela, S, Holger, W, Albert, B, Cuiying, Y, Xiaojing, C, Han, S, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-19 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand recognition and activation of neuromedin U receptor 2.
Nat Commun, 13, 2022
|
|
4BPM
 
 | | Crystal structure of a human integral membrane enzyme | | Descriptor: | 2-[[2,6-bis(chloranyl)-3-[(2,2-dimethylpropanoylamino)methyl]phenyl]amino]-1-methyl-6-(2-methyl-2-oxidanyl-propoxy)-N-[2,2,2-tris(fluoranyl)ethyl]benzimidazole-5-carboxamide, GLUTATHIONE, PROSTAGLANDIN E SYNTHASE, ... | | Authors: | Li, D, Wang, M, Olieric, V, Caffrey, M. | | Deposit date: | 2013-05-27 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystallizing Membrane Proteins in the Lipidic Mesophase. Experience with Human Prostaglandin E2 Synthase 1 and an Evolving Strategy.
Cryst.Growth Des., 14, 2014
|
|
4TN8
 
 | | Crystal structure of Thermus Thermophilus thioredoxin solved by sulfur SAD using Swiss Light Source data | | Descriptor: | CHLORIDE ION, Thioredoxin | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-06-03 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
