4A1X
 
 | | Co-Complex structure of NS3-4A protease with the inhibitory peptide CP5-46-A (Synchrotron data) | | Descriptor: | CHLORIDE ION, CP5-46-A PEPTIDE, NONSTRUCTURAL PROTEIN 4A, ... | | Authors: | Schmelz, S, Kuegler, J, Collins, J, Heinz, D. | | Deposit date: | 2011-09-20 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High Affinity Peptide Inhibitors of the Hepatitis C Virus Ns3-4A Protease Refractory to Common Resistant Mutants.
J.Biol.Chem., 287, 2012
|
|
4A1T
 
 | | Co-Complex of the of NS3-4A protease with the inhibitory peptide CP5- 46-A (in-House data) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BICARBONATE ION, CHLORIDE ION, ... | | Authors: | Schmelz, S, Kuegler, J, Collins, J, Heinz, D.W. | | Deposit date: | 2011-09-19 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | High Affinity Peptide Inhibitors of the Hepatitis C Virus Ns3-4A Protease Refractory to Common Resistant Mutants.
J.Biol.Chem., 287, 2012
|
|
4A1V
 
 | | Co-Complex structure of NS3-4A protease with the optimized inhibitory peptide CP5-46A-4D5E | | Descriptor: | CHLORIDE ION, CP5-46A-4D5E, NON-STRUCTURAL PROTEIN 4A, ... | | Authors: | Schmelz, S, Kuegler, J, Collins, J, Heinz, D.W. | | Deposit date: | 2011-09-20 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High Affinity Peptide Inhibitors of the Hepatitis C Virus Ns3-4A Protease Refractory to Common Resistant Mutants.
J.Biol.Chem., 287, 2012
|
|
4AOQ
 
 | | Cationic trypsin in complex with mutated Spinacia oleracea trypsin inhibitor III (SOTI-III) (F14A) | | Descriptor: | CALCIUM ION, CATIONIC TRYPSIN, PENTAETHYLENE GLYCOL, ... | | Authors: | Schmelz, S, Glotzbach, B, Reinwarth, M, Christmann, A, Kolmar, H, Heinz, D.W. | | Deposit date: | 2012-03-29 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of Spinacia Oleracea Trypsin Inhibitor III (Soti-III)
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4AOR
 
 | | Cationic trypsin in complex with the Spinacia oleracea trypsin inhibitor III (SOTI-III) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Schmelz, S, Glotzbach, B, Reinwarth, M, Christmann, A, Kolmar, H, Heinz, D.W. | | Deposit date: | 2012-03-29 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structural Characterization of Spinacia Oleracea Trypsin Inhibitor III (Soti-III)
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4ABI
 
 | | Co-complex structure of bovine trypsin with a modified Bowman-Birk inhibitor (PtA)SFTI-1(1,14), that was 1,4-disubstituted with a 1,2,3- trizol to mimic a trans amide bond | | Descriptor: | CALCIUM ION, CATIONIC TRYPSIN, DIMETHYLFORMAMIDE, ... | | Authors: | Schmelz, S, Empting, M, Tischler, M, Nasu, D, Heinz, D, Kolmar, H. | | Deposit date: | 2011-12-08 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Braces for the Peptide Backbone: Insights Into Structure-Activity Relation-Ships of Protease Inhibitor Mimics with Locked Amide Conformations
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4ABJ
 
 | | Co-complex structure of bovine trypsin with a modified Bowman-Birk inhibitor (IcA)SFTI-1(1,14), that was 1,5-disubstituted with 1,2,3- trizol to mimic a cis amide bond | | Descriptor: | CALCIUM ION, CATIONIC TRYPSIN, DIMETHYLFORMAMIDE, ... | | Authors: | Schmelz, S, Empting, M, Tischler, M, Nasu, D, Heinz, D, Kolmar, H. | | Deposit date: | 2011-12-08 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Braces for the Peptide Backbone: Insights Into Structure-Activity Relation-Ships of Protease Inhibitor Mimics with Locked Amide Conformations
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4B0F
 
 | | Heptameric core complex structure of C4b-binding (C4BP) protein from human | | Descriptor: | C4B-BINDING PROTEIN ALPHA CHAIN, CHLORIDE ION | | Authors: | Schmelz, S, Hofmeyer, T, Kolmar, H, Heinz, D.W. | | Deposit date: | 2012-07-02 | | Release date: | 2013-01-09 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Arranged Sevenfold: Structural Insights Into the C-Terminal Oligomerization Domain of Human C4B-Binding Protein.
J.Mol.Biol., 425, 2013
|
|
4CH7
 
 | | Crystal structure of the siroheme decarboxylase NirDL | | Descriptor: | NIRD-LIKE PROTEIN | | Authors: | Schmelz, S, Kriegler, T.M, Haufschildt, K, Layer, G, Heinz, D.W. | | Deposit date: | 2013-11-29 | | Release date: | 2014-07-30 | | Last modified: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | The Crystal Structure of Siroheme Decarboxylase in Complex with Iron-Uroporphyrin III Reveals Two Essential Histidine Residues
J.Mol.Biol., 426, 2014
|
|
4CZC
 
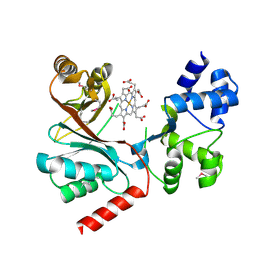 | | Crystal structure of the siroheme decarboxylase NirDL in co-complex with iron-uroporphyrin III analogue | | Descriptor: | Fe(III) Uroporphyrin, NIRD-LIKE PROTEIN | | Authors: | Schmelz, S, Kriegler, T.M, Haufschildt, K, Layer, G, Heinz, D.W. | | Deposit date: | 2014-04-17 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Crystal Structure of Siroheme Decarboxylase in Complex with Iron-Uroporphyrin III Reveals Two Essential Histidine Residues
J.Mol.Biol., 426, 2014
|
|
7R3J
 
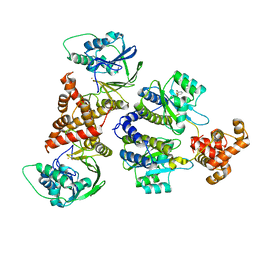 | | Nativ complex of PqsE and RhlR with the synthetic antagonist mBTL | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide, FE (III) ION, ... | | Authors: | Borgert, S.R, Schmelz, S, Blankenfeldt, W. | | Deposit date: | 2022-02-07 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
7R3F
 
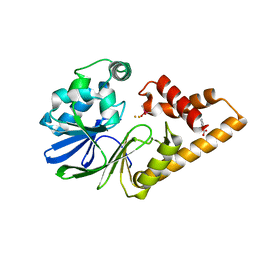 | | Monomeric PqsE mutant E187R | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, BENZOIC ACID, CACODYLATE ION, ... | | Authors: | Borgert, S.R, Schmelz, S, Blankenfeldt, W. | | Deposit date: | 2022-02-07 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
7R3E
 
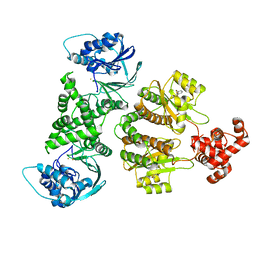 | | Fusion construct of PqsE and RhlR in complex with the synthetic antagonist mBTL | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase,Regulatory protein RhlR, 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide, FE (III) ION | | Authors: | Borgert, S.R, Schmelz, S, Blankenfeldt, W. | | Deposit date: | 2022-02-07 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
2X0O
 
 | | Apo structure of the Alcaligin biosynthesis protein C (AlcC) from Bordetella bronchiseptica | | Descriptor: | ALCALIGIN BIOSYNTHESIS PROTEIN, SULFATE ION | | Authors: | Johnson, K.A, Schmelz, S, Kadi, N, Mcmahon, S.A, Oke, M, Liu, H, Carter, L.G, White, M.F, Challis, G.L, Naismith, J.H. | | Deposit date: | 2009-12-16 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
8B4A
 
 | | Nativ complex of PqsE and RhlR with autoinducer C4-HSL | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, FE (III) ION, N-[(3S)-2-oxotetrahydrofuran-3-yl]butanamide, ... | | Authors: | Borgert, S.R, Blankenfeldt, W. | | Deposit date: | 2022-09-20 | | Release date: | 2022-12-14 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
6Q7U
 
 | |
6Q7W
 
 | |
6Q7V
 
 | |
7R3I
 
 | |
7R3G
 
 | |
7R3H
 
 | |
2IVY
 
 | | Crystal structure of hypothetical protein sso1404 from Sulfolobus solfataricus P2 | | Descriptor: | HYPOTHETICAL PROTEIN SSO1404 | | Authors: | Yan, X, Carter, L.G, Dorward, M, Liu, H, McMahon, S.A, Oke, M, Powers, H, White, M.F, Naismith, J.H. | | Deposit date: | 2006-06-22 | | Release date: | 2006-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2JG5
 
 | | CRYSTAL STRUCTURE OF A PUTATIVE PHOSPHOFRUCTOKINASE FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | FRUCTOSE 1-PHOSPHATE KINASE | | Authors: | Yan, X, Carter, L.G, Johnson, K.A, Liu, H, Dorward, M, McMahon, S.A, Oke, M, Powers, H, Coote, P.J, Naismith, J.H. | | Deposit date: | 2007-02-08 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2JG6
 
 | | CRYSTAL STRUCTURE OF A 3-METHYLADENINE DNA GLYCOSYLASE I FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | DNA-3-METHYLADENINE GLYCOSIDASE, ZINC ION | | Authors: | Yan, X, Carter, L.G, Liu, H, Dorward, M, McMahon, S.A, Johnson, K.A, Oke, M, Coote, P.J, Naismith, J.H. | | Deposit date: | 2007-02-08 | | Release date: | 2007-02-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X48
 
 | | ORF 55 from Sulfolobus islandicus rudivirus 1 | | Descriptor: | CAG38821, PHOSPHATE ION | | Authors: | Oke, M, Carter, L, Johnson, K.A, Liu, H, Mcmahon, S, Naismith, J.H, White, M.F. | | Deposit date: | 2010-01-28 | | Release date: | 2010-07-21 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
