6BP2
 
 | | Therapeutic human monoclonal antibody MR191 bound to a marburgvirus glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, Envelope glycoprotein GP2, ... | | Authors: | King, L.B, Fusco, M.L, Flyak, A.I, Ilinykh, P.A, Huang, K, Gunn, B, Kirchdoerfer, R.N, Hastie, K.M, Sangha, A.K, Meiler, J, Alter, G, Bukreyev, A, Crowe, J.E.J, Saphire, E.O. | | Deposit date: | 2017-11-21 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.172 Å) | | Cite: | The Marburgvirus-Neutralizing Human Monoclonal Antibody MR191 Targets a Conserved Site to Block Virus Receptor Binding.
Cell Host Microbe, 23, 2018
|
|
5WEQ
 
 | | The crystal structure of a MR78 mutant | | Descriptor: | MR78 mutant Fab heavy chain, MR78 mutant light chain | | Authors: | Dong, J, Williamson, L.E, Crowe, J.E. | | Deposit date: | 2017-07-10 | | Release date: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of Non-local Interactions between CDR Loops in Binding Affinity of MR78 Antibody to Marburg Virus Glycoprotein.
Structure, 25, 2017
|
|
5JRP
 
 | | crystal structure of monoclonal antibody MR78 Fab | | Descriptor: | SODIUM ION, marberg virus monoclonal antibody MR78 Fab heavy chain, marberg virus monoclonal antibody MR78 Fab light chain | | Authors: | Dong, J, Crowe, J. | | Deposit date: | 2016-05-06 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of Non-local Interactions between CDR Loops in Binding Affinity of MR78 Antibody to Marburg Virus Glycoprotein.
Structure, 25, 2017
|
|
6V4R
 
 | |
6V4Q
 
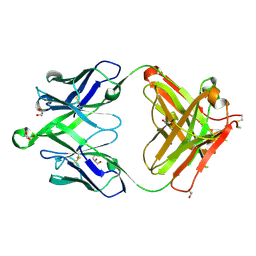 | | Crystal structure of a MR78-like antibody naive-1 Fab | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, Naive-1 Fab heavy chain, ... | | Authors: | Bozhanova, N.G, Crowe, J.E, Meiler, J. | | Deposit date: | 2019-11-28 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Discovery of Marburg virus neutralizing antibodies from virus-naive human antibody repertoires using large-scale structural predictions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
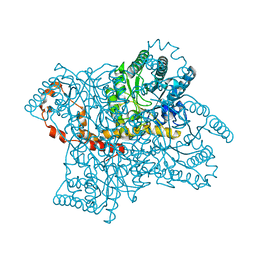
4QEE
 
 | |
4QDW
 
 | | Joint X-ray and neutron structure of Streptomyces rubiginosus D-xylose isomerase in complex with two Ni2+ ions and linear L-arabinose | | Descriptor: | L-arabinose, NICKEL (II) ION, Xylose isomerase | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2014-05-14 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | Cite: | L-Arabinose Binding, Isomerization, and Epimerization by D-Xylose Isomerase: X-Ray/Neutron Crystallographic and Molecular Simulation Study.
Structure, 22, 2014
|
|
4QE5
 
 | |
4QE4
 
 | |
4QDP
 
 | | Joint X-ray and neutron structure of Streptomyces rubiginosus D-xylose isomerase in complex with two Cd2+ ions and cyclic beta-L-arabinose | | Descriptor: | CADMIUM ION, Xylose isomerase, beta-L-arabinopyranose | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2014-05-14 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | L-Arabinose Binding, Isomerization, and Epimerization by D-Xylose Isomerase: X-Ray/Neutron Crystallographic and Molecular Simulation Study.
Structure, 22, 2014
|
|
4QEH
 
 | |
4QE1
 
 | |
