2PZS
 
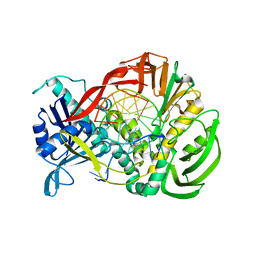 | | Phi29 DNA polymerase complexed with primer-template DNA (post-translocation binary complex) | | Descriptor: | 5'-d(CTAACACGTAAGCAGTC)-3', 5'-d(GACTGCTTAC)-3', DNA polymerase | | Authors: | Berman, A.J, Kamtekar, S, Goodman, J.L, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2007-05-18 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases
Embo J., 26, 2007
|
|
1ZAE
 
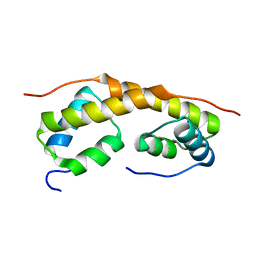 | | Solution structure of the functional domain of phi29 replication organizer p16.7c | | Descriptor: | Early protein GP16.7 | | Authors: | Asensio, J.L, Albert, A, Munoz-Espin, D, Gonzalez, C, Hermoso, J, Villar, L, Jimenez-Barbero, J, Salas, M, Meijer, W.J.J. | | Deposit date: | 2005-04-06 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the functional domain of phi29 replication organizer: insights into oligomerization and dna binding
J.Biol.Chem., 280, 2005
|
|
2LE2
 
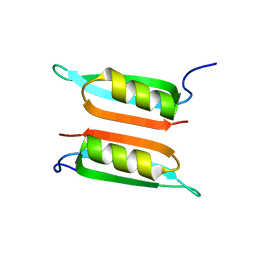 | | Novel dimeric structure of phage phi29-encoded protein p56: Insights into Uracil-DNA glycosylase inhibition | | Descriptor: | P56 | | Authors: | Asensio, J, Perez-Lago, L, Lazaro, J.M, Gonzalez, C, Serrano-Heras, G, Salas, M. | | Deposit date: | 2011-06-06 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Novel dimeric structure of phage 29-encoded protein p56: insights into uracil-DNA glycosylase inhibition.
Nucleic Acids Res., 39, 2011
|
|
3ZOR
 
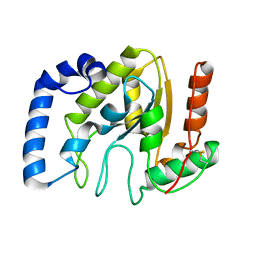 | | Structure of BsUDG | | Descriptor: | URACIL-DNA GLYCOSYLASE | | Authors: | Banos-Sanz, J.I, Mojardin, L, Sanz-Aparicio, J, Gonzalez, B, Salas, M. | | Deposit date: | 2013-02-22 | | Release date: | 2013-05-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure and Functional Insights Into Uracil-DNA Glycosylase Inhibition by Phage Phi29 DNA Mimic Protein P56
Nucleic Acids Res., 41, 2013
|
|
3ZOQ
 
 | | Structure of BsUDG-p56 complex | | Descriptor: | CHLORIDE ION, GLYCEROL, P56, ... | | Authors: | Banos-Sanz, J.I, Mojardin, L, Sanz-Aparicio, J, Gonzalez, B, Salas, M. | | Deposit date: | 2013-02-22 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure and Functional Insights Into Uracil-DNA Glycosylase Inhibition by Phage Phi29 DNA Mimic Protein P56
Nucleic Acids Res., 41, 2013
|
|
2PY5
 
 | | Phi29 DNA polymerase complexed with single-stranded DNA | | Descriptor: | 1,2-ETHANEDIOL, 5'-d(GGACTTT)-3', DNA polymerase | | Authors: | Berman, A.J, Kamtekar, S, Goodman, J.L, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2007-05-15 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases
Embo J., 26, 2007
|
|
2C5R
 
 | | The structure of phage phi29 replication organizer protein p16.7 in complex with double stranded DNA | | Descriptor: | 5'-D(*CP*CP*GP*GP*TP*GP*GP*AP)-3', 5'-D(*TP*CP*CP*AP*CP*CP*GP*GP)-3', EARLY PROTEIN P16.7 | | Authors: | Albert, A, Jimenez, M, Munoz-Espin, D, Asensio, J.L, Hermoso, J.A, Salas, M, Meijer, W.J.J. | | Deposit date: | 2005-10-31 | | Release date: | 2005-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for Membrane Anchorage of Viral Phi 29 DNA During Replication.
J.Biol.Chem., 280, 2005
|
|
1XHX
 
 | | Phi29 DNA Polymerase, orthorhombic crystal form | | Descriptor: | DNA polymerase, MAGNESIUM ION, SULFATE ION | | Authors: | Kamtekar, S, Berman, A.J, Wang, J, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2004-09-21 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Insights into Strand Displacement and Processivity from the Crystal Structure of the Protein-Primed DNA Polymerase of Bacteriophage phi29
Mol.Cell, 16, 2004
|
|
1XHZ
 
 | | Phi29 DNA polymerase, orthorhombic crystal form, ssDNA complex | | Descriptor: | 5'-D(*TP*TP*TP*TP*T)-3', DNA polymerase | | Authors: | Kamtekar, S, Berman, A.J, Wang, J, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2004-09-21 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into Strand Displacement and Processivity from the Crystal Structure of the Protein-Primed DNA Polymerase of Bacteriophage phi29
Mol.Cell, 16, 2004
|
|
2PYL
 
 | | Phi29 DNA polymerase complexed with primer-template DNA and incoming nucleotide substrates (ternary complex) | | Descriptor: | 1,2-ETHANEDIOL, 5'-d(CTGACGAATGTACA)-3', 5'-d(GACTGCTTAC(2DA))-3', ... | | Authors: | Berman, A.J, Kamtekar, S, Goodman, J.L, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2007-05-16 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases
Embo J., 26, 2007
|
|
2PYJ
 
 | | Phi29 DNA polymerase complexed with primer-template DNA and incoming nucleotide substrates (ternary complex) | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-d(ACACGTAAGCAGTC)-3', ... | | Authors: | Berman, A.J, Kamtekar, S, Goodman, J.L, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2007-05-16 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases
Embo J., 26, 2007
|
|
2FIO
 
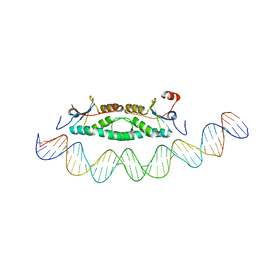 | | Phage phi29 transcription regulator p4-DNA complex | | Descriptor: | DNA (41-MER), Late genes activator | | Authors: | Badia, D, Camacho, A, Perez-Lago, L, Escandon, C, Salas, M, Coll, M. | | Deposit date: | 2005-12-30 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of phage phi29 transcription regulator p4-DNA complex reveals an N-hook motif for DNA
Mol.Cell, 22, 2006
|
|
2FIP
 
 | | Phage phi29 transcription regulator p4 | | Descriptor: | Late genes activator | | Authors: | Badia, D, Camacho, A, Perez-Lago, L, Escandon, C, Salas, M, Coll, M. | | Deposit date: | 2005-12-30 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of phage phi29 transcription regulator p4-DNA complex reveals an N-hook motif for DNA
Mol.Cell, 22, 2006
|
|
2EX3
 
 | | Bacteriophage phi29 DNA polymerase bound to terminal protein | | Descriptor: | DNA polymerase, DNA terminal protein, LEAD (II) ION | | Authors: | Kamtekar, S, Berman, A.J, Wang, J, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2005-11-07 | | Release date: | 2006-03-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The phi29 DNA polymerase:protein-primer structure suggests a model for the initiation to elongation transition
Embo J., 25, 2006
|
|
2BNK
 
 | | The structure of phage phi29 replication organizer protein p16.7 | | Descriptor: | EARLY PROTEIN GP16.7 | | Authors: | Albert, A, Asensio, J.L, Munoz-Espin, D, Gonzalez, C, Hermoso, J.A, Villar, L, Jimenez-Barbero, J, Salas, M, Meijer, W.J.J. | | Deposit date: | 2005-03-28 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Functional Domain of {Varphi}29 Replication Organizer: Insights Into Oligomerization and DNA Binding.
J.Biol.Chem., 280, 2005
|
|
3CLE
 
 | |
3CLF
 
 | |
1XI1
 
 | | Phi29 DNA polymerase ssDNA complex, monoclinic crystal form | | Descriptor: | 5'-D(P*TP*TP*TP*TP*T)-3', DNA polymerase, MAGNESIUM ION | | Authors: | Kamtekar, S, Berman, A.J, Wang, J, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2004-09-21 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Correction of X-ray intensities from single crystals containing lattice-translocation defects
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4WAG
 
 | | Phosphatidylinositol 4-kinase III beta crystallized with MI103 inhibitor | | Descriptor: | 6-chloro-3-(3,4-dimethoxyphenyl)-2-methylimidazo[1,2-b]pyridazin-8-amine, Phosphatidylinositol 4-kinase beta,Phosphatidylinositol 4-kinase beta | | Authors: | Chalupska, D, Boura, E. | | Deposit date: | 2014-08-29 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.407 Å) | | Cite: | Highly Selective Phosphatidylinositol 4-Kinase III beta Inhibitors and Structural Insight into Their Mode of Action.
J.Med.Chem., 58, 2015
|
|
4WAE
 
 | | Phosphatidylinositol 4-kinase III beta crystallized with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Phosphatidylinositol 4-kinase beta,Phosphatidylinositol 4-kinase beta | | Authors: | Chalupska, D, Boura, E. | | Deposit date: | 2014-08-29 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.318 Å) | | Cite: | Highly Selective Phosphatidylinositol 4-Kinase III beta Inhibitors and Structural Insight into Their Mode of Action.
J.Med.Chem., 58, 2015
|
|
7Q3B
 
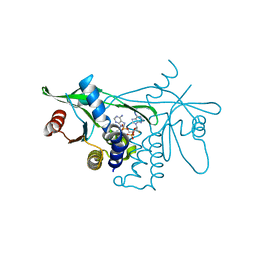 | | Crystal structure of human STING in complex with 3'3'-c-(2'F,2'dA-isonucA)MP | | Descriptor: | 3'3'-c-(2'F,2'dA-isonucA)MP, Stimulator of interferon genes protein | | Authors: | Smola, M, Klima, M, Boura, E. | | Deposit date: | 2021-10-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.55733919 Å) | | Cite: | Discovery of isonucleotidic CDNs as potent STING agonists with immunomodulatory potential.
Structure, 30, 2022
|
|
8ORW
 
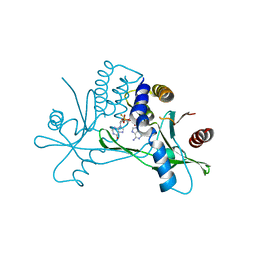 | | Crystal structure of human STING in complex with the agonist MD1203 | | Descriptor: | 9-[(1~{S},6~{R},8~{R},9~{R},10~{R},15~{R},17~{R},18~{R})-8-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, Stimulator of interferon protein | | Authors: | Klima, M, Boura, E. | | Deposit date: | 2023-04-17 | | Release date: | 2023-12-20 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Fluorinated cGAMP analogs, which act as STING agonists and are not cleavable by poxins: Structural basis of their function.
Structure, 32, 2024
|
|
8ORV
 
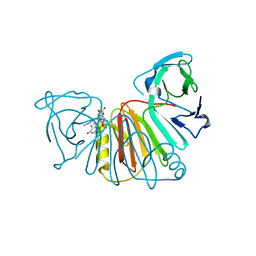 | | Crystal structure of monkeypox virus poxin in complex with the STING agonist MD1203 | | Descriptor: | 9-[(1~{S},6~{R},8~{R},9~{R},10~{R},15~{R},17~{R},18~{R})-8-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, MPXVgp165 | | Authors: | Duchoslav, V, Klima, M, Boura, E. | | Deposit date: | 2023-04-17 | | Release date: | 2023-12-20 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Fluorinated cGAMP analogs, which act as STING agonists and are not cleavable by poxins: Structural basis of their function.
Structure, 32, 2024
|
|
4Y8D
 
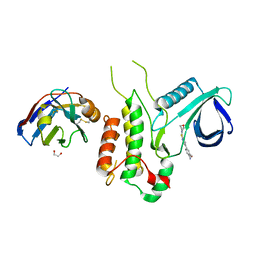 | | Crystal structure of Cyclin-G associated kinase (GAK) complexed with selective 12i inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-methoxy-4-[3-(morpholin-4-yl)[1,2]thiazolo[4,3-b]pyridin-6-yl]aniline, Cyclin-G-associated kinase, ... | | Authors: | Chaikuad, A, Heroven, C, Nowak, R, De Jonghe, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-02-16 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Selective Inhibitors of Cyclin G Associated Kinase (GAK) as Anti-Hepatitis C Agents.
J.Med.Chem., 58, 2015
|
|
8P45
 
 | | Crystal structure of human STING in complex with the agonist MD1202D | | Descriptor: | 9-[(1~{S},3~{R},6~{R},8~{R},9~{R},10~{R},12~{R},15~{R},17~{R},18~{R})-8-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanylidene)-3,12-bis(sulfanyl)-2,4,7,11,13-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, Stimulator of interferon genes protein | | Authors: | Klima, M, Boura, E. | | Deposit date: | 2023-05-19 | | Release date: | 2023-12-20 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Fluorinated cGAMP analogs, which act as STING agonists and are not cleavable by poxins: Structural basis of their function.
Structure, 32, 2024
|
|
