7T4Q
 
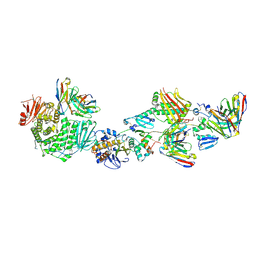 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with neutralizing fabs 2C12, 7I13 and 13H11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, Envelope glycoprotein L, ... | | Authors: | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
7T4R
 
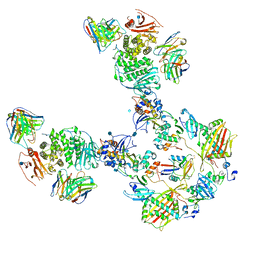 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with THBD and neutralizing fabs MSL-109 and 13H11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, Envelope glycoprotein L, ... | | Authors: | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
7T4S
 
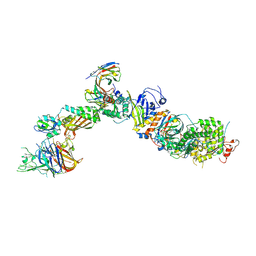 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with NRP2 and neutralizing fabs 8I21 and 13H11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
8F0Q
 
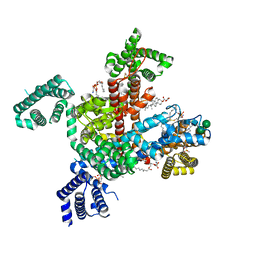 | | Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the acylsulfonamide inhibitor GDC-0310 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-cyclopropyl-4-({1-[(1S)-1-(3,5-dichlorophenyl)ethyl]piperidin-4-yl}methoxy)-2-fluoro-N-(methanesulfonyl)benzamide, ... | | Authors: | Kschonsak, M, Jao, C.C, Arthur, C.P, Rohou, A.L, Bergeron, P, Ortwine, D, McKerall, S.J, Hackos, D.H, Deng, L, Chen, J, Sutherlin, D, Dragovich, P.S, Volgraf, M, Wright, M.R, Payandeh, J, Ciferri, C, Tellis, J.C. | | Deposit date: | 2022-11-03 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM reveals an unprecedented binding site for Na V 1.7 inhibitors enabling rational design of potent hybrid inhibitors.
Elife, 12, 2023
|
|
8F0P
 
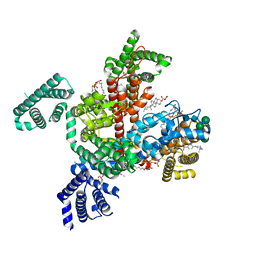 | | Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the hybrid inhibitor GNE-1305 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Kschonsak, M, Jao, C.C, Arthur, C.P, Rohou, A.L, Bergeron, P, Ortwine, D, McKerall, S.J, Hackos, D.H, Deng, L, Chen, J, Sutherlin, D, Dragovich, P.S, Volgraf, M, Wright, M.R, Payandeh, J, Ciferri, C, Tellis, J.C. | | Deposit date: | 2022-11-03 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Cryo-EM reveals an unprecedented binding site for Na V 1.7 inhibitors enabling rational design of potent hybrid inhibitors.
Elife, 12, 2023
|
|
8F0S
 
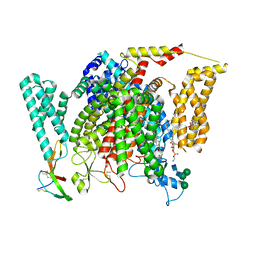 | | Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the hybrid inhibitor GNE-9296 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kschonsak, M, Jao, C.C, Arthur, C.P, Rohou, A.L, Bergeron, P, Ortwine, D, McKerall, S.J, Hackos, D.H, Deng, L, Chen, J, Sutherlin, D, Dragovich, P.S, Volgraf, M, Wright, M.R, Payandeh, J, Ciferri, C, Tellis, J.C. | | Deposit date: | 2022-11-03 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM reveals an unprecedented binding site for Na V 1.7 inhibitors enabling rational design of potent hybrid inhibitors.
Elife, 12, 2023
|
|
8F0R
 
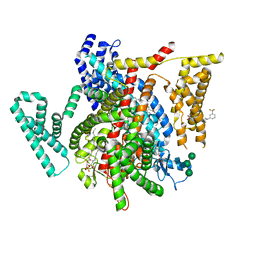 | | Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the arylsulfonamide inhibitor GNE-3565 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-chloro-4-({(1S,2S,4S)-2-(dimethylamino)-4-[3-(trifluoromethyl)phenyl]cyclohexyl}amino)-2-fluoro-N-(pyrimidin-4-yl)benzene-1-sulfonamide, ... | | Authors: | Kschonsak, M, Jao, C.C, Arthur, C.P, Rohou, A.L, Bergeron, P, Ortwine, D, McKerall, S.J, Hackos, D.H, Deng, L, Chen, J, Sutherlin, D, Dragovich, P.S, Volgraf, M, Wright, M.R, Payandeh, J, Ciferri, C, Tellis, J.C. | | Deposit date: | 2022-11-03 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM reveals an unprecedented binding site for Na V 1.7 inhibitors enabling rational design of potent hybrid inhibitors.
Elife, 12, 2023
|
|
7LBE
 
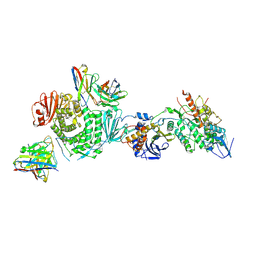 | | CryoEM structure of the HCMV Trimer gHgLgO in complex with neutralizing fabs 13H11 and MSL-109 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Rouge, L, Arthur, C.P, Hoangdung, H, Patel, N, Kim, I, Johnson, M, Kraft, E, Rohou, A.L, Gill, A, Martinez-Martin, N, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-10 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry.
Cell, 184, 2021
|
|
7LBG
 
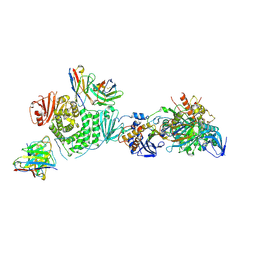 | | CryoEM structure of the HCMV Trimer gHgLgO in complex with human Transforming growth factor beta receptor type 3 and neutralizing fabs 13H11 and MSL-109 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Rouge, L, Arthur, C.P, Hoangdung, H, Patel, N, Kim, I, Johnson, M, Kraft, E, Rohou, A.L, Gill, A, Martinez-Martin, N, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-10 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry.
Cell, 184, 2021
|
|
7LBF
 
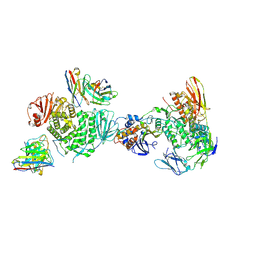 | | CryoEM structure of the HCMV Trimer gHgLgO in complex with human Platelet-derived growth factor receptor alpha and neutralizing fabs 13H11 and MSL-109 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Rouge, L, Arthur, C.P, Hoangdung, H, Patel, N, Kim, I, Johnson, M, Kraft, E, Rohou, A.L, Gill, A, Martinez-Martin, N, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-10 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry.
Cell, 184, 2021
|
|
