4XJE
 
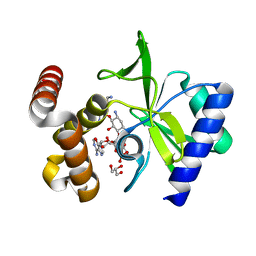 | |
5CFS
 
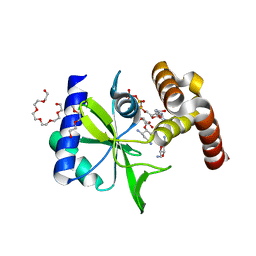 | | Crystal Structure of ANT(2")-Ia in complex with AMPCPP and tobramycin | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, AAD(2''),Gentamicin 2''-nucleotidyltransferase,Gentamicin resistance protein, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Rodionov, D, Bassenden, A.V, Berghuis, A.M. | | Deposit date: | 2015-07-08 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Analysis of the Tobramycin and Gentamicin Clinical Resistome Reveals Limitations for Next-generation Aminoglycoside Design.
Acs Chem.Biol., 11, 2016
|
|
5CFT
 
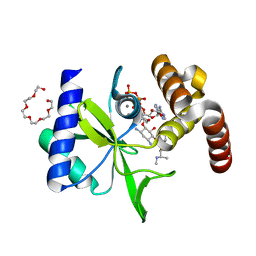 | | Crystal Structure of ANT(2")-Ia in complex with AMPCPP and gentamicin C1 | | Descriptor: | Aminoglycoside Nucleotidyltransferase (2")-Ia, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, HEXAETHYLENE GLYCOL, ... | | Authors: | Rodionov, D, Bassenden, A.V, Berghuis, A.M. | | Deposit date: | 2015-07-08 | | Release date: | 2016-03-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis of the Tobramycin and Gentamicin Clinical Resistome Reveals Limitations for Next-generation Aminoglycoside Design.
Acs Chem.Biol., 11, 2016
|
|
5CFU
 
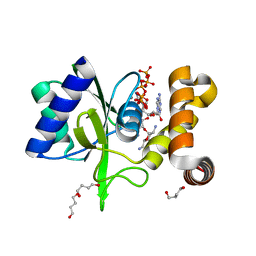 | | Crystal Structure of ANT(2")-Ia in complex with adenylyl-2"-tobramycin | | Descriptor: | 1,4-BUTANEDIOL, Aminoglycoside Nucleotidyltransferase (2")-Ia, MANGANESE (II) ION, ... | | Authors: | Rodionov, D, Bassenden, A.V, Berghuis, A.M. | | Deposit date: | 2015-07-08 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Revisiting the Catalytic Cycle and Kinetic Mechanism of AminoglycosideO-Nucleotidyltransferase(2′′): A Structural and Kinetic Study.
Acs Chem.Biol., 2020
|
|
4PVX
 
 | | Crystal structure of human FPPS in complex with [({4-[4-(cyclopropyloxy)phenyl]pyridin-2-yl}amino)methanediyl]bis(phosphonic acid) | | Descriptor: | Farnesyl pyrophosphate synthase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Rodionov, D, Park, J, Lin, Y.-S, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2014-03-18 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystallographic and thermodynamic characterization of phenylaminopyridine bisphosphonates binding to human farnesyl pyrophosphate synthase.
PLoS ONE, 12, 2017
|
|
4PVY
 
 | | Crystal structure of human FPPS in complex with [({5-[4-(propan-2-yloxy)phenyl]pyridin-3-yl}amino)methanediyl]bis(phosphonic acid) | | Descriptor: | Farnesyl pyrophosphate synthase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Rodionov, D, Park, J, De Schutter, J.W, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2014-03-18 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic and thermodynamic characterization of phenylaminopyridine bisphosphonates binding to human farnesyl pyrophosphate synthase.
PLoS ONE, 12, 2017
|
|
4JRD
 
 | | Crystal structure of the parallel double-stranded helix of poly(A) RNA | | Descriptor: | AMMONIUM ION, RNA (5'-R(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3') | | Authors: | Safaee, N, Noronha, A.M, Kozlov, G, Rodionov, D, Wilds, C.J, Sheldrick, G.M, Gehring, K. | | Deposit date: | 2013-03-21 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure of the parallel duplex of poly(A) RNA: evaluation of a 50 year-old prediction.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
5DD4
 
 | | Apo structure of transcriptional factor AraR from Bacteroides thetaiotaomicron VPI | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, TRANSCRIPTIONAL REGULATOR AraR | | Authors: | Chang, C, Tesar, C, Rodionov, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-08-24 | | Release date: | 2015-09-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | A novel transcriptional regulator of L-arabinose utilization in human gut bacteria.
Nucleic Acids Res., 43, 2015
|
|
5DDG
 
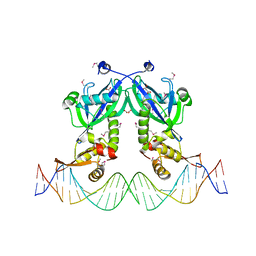 | | The structure of transcriptional factor AraR from Bacteroides thetaiotaomicron VPI in complex with target double strand DNA | | Descriptor: | DNA (27-MER), FORMIC ACID, MALONIC ACID, ... | | Authors: | Chang, C, Tesar, C, Rodionov, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-08-24 | | Release date: | 2015-09-09 | | Last modified: | 2015-12-16 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | A novel transcriptional regulator of L-arabinose utilization in human gut bacteria.
Nucleic Acids Res., 43, 2015
|
|
5DEQ
 
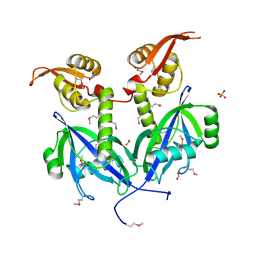 | | Crystal structure of transcriptional factor AraR from Bacteroides thetaiotaomicron VPI in complex with L-arabinose | | Descriptor: | FORMIC ACID, SULFATE ION, TRANSCRIPTIONAL REGULATOR AraR, ... | | Authors: | Chang, C, Tesar, C, Rodionov, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-08-25 | | Release date: | 2015-10-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A novel transcriptional regulator of L-arabinose utilization in human gut bacteria.
Nucleic Acids Res., 43, 2015
|
|
1ZKG
 
 | |
4JN3
 
 | |
4JN5
 
 | |
4ZYN
 
 | | Crystal Structure of Parkin E3 ubiquitin ligase (linker deletion; delta 86-130) | | Descriptor: | E3 ubiquitin-protein ligase parkin, SULFATE ION, ZINC ION | | Authors: | Lilov, A, Sauve, V, Trempe, J.F, Rodionov, D, Wang, J, Gehring, K. | | Deposit date: | 2015-05-21 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | A Ubl/ubiquitin switch in the activation of Parkin.
Embo J., 34, 2015
|
|
4NFI
 
 | | Crystal structure of human FPPS in complex with magnesium and JDS05120 | | Descriptor: | Farnesyl pyrophosphate synthase, MAGNESIUM ION, [({5-[4-(cyclopropyloxy)phenyl]pyridin-3-yl}amino)methanediyl]bis(phosphonic acid) | | Authors: | Park, J, De Schutter, J.W, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2013-10-31 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic and thermodynamic characterization of phenylaminopyridine bisphosphonates binding to human farnesyl pyrophosphate synthase.
PLoS ONE, 12, 2017
|
|
4NFJ
 
 | | Crystal structure of human FPPS in complex with magnesium, JDS05120, and sulfate | | Descriptor: | Farnesyl pyrophosphate synthase, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Park, J, De Schutter, J.W, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2013-10-31 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic and thermodynamic characterization of phenylaminopyridine bisphosphonates binding to human farnesyl pyrophosphate synthase.
PLoS ONE, 12, 2017
|
|
4NFK
 
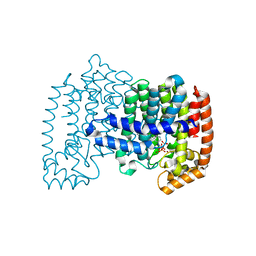 | | Crystal structure of human FPPS in complex with nickel, JDS05120, and sulfate | | Descriptor: | Farnesyl pyrophosphate synthase, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Park, J, De schutter, J.W, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2013-10-31 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic and thermodynamic characterization of phenylaminopyridine bisphosphonates binding to human farnesyl pyrophosphate synthase.
PLoS ONE, 12, 2017
|
|
