7BAW
 
 | | A heptameric barrel state of a de novo coiled-coil assembly: CC-Type2-(GgIaId)4 | | Descriptor: | CC-Type2-(GgIaId)4, GLYCEROL, N-PROPANOL | | Authors: | Martin, F.J.O, Dawson, W.M, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2020-12-16 | | Release date: | 2021-04-28 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Coiled coils 9-to-5: rational de novo design of alpha-helical barrels with tunable oligomeric states.
Chem Sci, 12, 2021
|
|
7BO9
 
 | |
7BO8
 
 | | A hexameric de novo coiled-coil assembly: CC-Type2-(VaYd)4-Y3F-W19(BrPhe)-Y24F. | | Descriptor: | 1,2-ETHANEDIOL, CC-Type2-(VaYd)4-Y3F-W19(BrPhe)-Y24F, OXAMIC ACID | | Authors: | Rhys, G.G, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2021-01-24 | | Release date: | 2021-05-19 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | How Coiled-Coil Assemblies Accommodate Multiple Aromatic Residues.
Biomacromolecules, 22, 2021
|
|
7BOA
 
 | |
7BLJ
 
 | |
7BLK
 
 | | Structure of CBM BT3015C from Bacteroides thetaiotaomicron | | Descriptor: | CALCIUM ION, family 32 carbohydrate-binding module from Bacteroides thetaiotaomicron | | Authors: | Costa, R.L. | | Deposit date: | 2021-01-18 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Structural basis for mucin-type O-glycan recognition by proteins of a Bacteroides thetaiotaomicron polysaccharide utilization loci
To Be Published
|
|
7BLL
 
 | |
7BLG
 
 | | Structure of CBM BT3015C from Bacteroides thetaiotaomicron in complex with galactose | | Descriptor: | CALCIUM ION, beta-D-galactopyranose, family 32 carbohydrate-binding module from Bacteroides thetaiotaomicron | | Authors: | Costa, R.L. | | Deposit date: | 2021-01-18 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structural basis for mucin-type O-glycan recognition by proteins of a Bacteroides thetaiotaomicron polysaccharide utilization loci
To Be Published
|
|
7BLH
 
 | |
1HCF
 
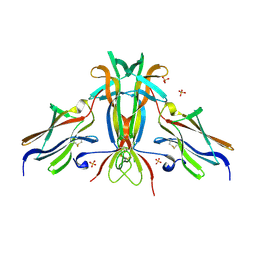 | | Crystal structure of TrkB-d5 bound to neurotrophin-4/5 | | Descriptor: | BDNF/NT-3 GROWTH FACTORS RECEPTOR, NEUROTROPHIN-4, SULFATE ION | | Authors: | Banfield, M.J, Naylor, R.L, Robertson, A.G.S, Allen, S.J, Dawbarn, D, Brady, R.L. | | Deposit date: | 2001-05-03 | | Release date: | 2001-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Specificity in Trk-Receptor:Neurotrophin Interaction: The Crystal Structure of Trkb-D5 in Complex with Neurotrophin-4/5
Structure, 9, 2001
|
|
5SXY
 
 | |
2NCK
 
 | |
1SMB
 
 | | Crystal Structure of Golgi-Associated PR-1 protein | | Descriptor: | 17kD fetal brain protein | | Authors: | Serrano, R.L, Kuhn, A, Hendricks, A, Helms, J.B, Sinning, I, Groves, M.R. | | Deposit date: | 2004-03-08 | | Release date: | 2004-09-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analysis of the human Golgi-associated plant pathogenesis related protein GAPR-1 implicates dimerization as a regulatory mechanism
J.Mol.Biol., 339, 2004
|
|
1T6O
 
 | | Nucleocapsid-binding domain of the measles virus P protein (amino acids 457-507) in complex with amino acids 486-505 of the measles virus N protein | | Descriptor: | linker, phosphoprotein | | Authors: | Kingston, R.L, Hamel, D.J, Gay, L.S, Dahlquist, F.W, Matthews, B.W. | | Deposit date: | 2004-05-06 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the attachment of a paramyxoviral polymerase to its template.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1T27
 
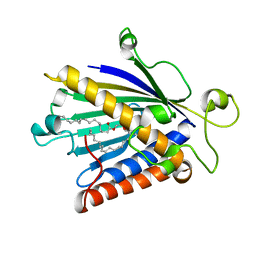 | | THE STRUCTURE OF PITP COMPLEXED TO PHOSPHATIDYLCHOLINE | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Phosphatidylinositol transfer protein alpha isoform | | Authors: | Yoder, M.D, Thomas, L.M, Tremblay, J.M, Oliver, R.L, Yarbrough, L.R, Helmkamp Jr, G.M. | | Deposit date: | 2004-04-20 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | STRUCTURE OF A MULTIFUNCTIONAL PROTEIN. MAMMALIAN PHOSPHATIDYLINOSITOL TRANSFER PROTEIN COMPLEXED WITH PHOSPHATIDYLCHOLINE
J.Biol.Chem., 276, 2001
|
|
6YRD
 
 | | SFX structure of dye-type peroxidase DtpB in the ferryl state | | Descriptor: | MAGNESIUM ION, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lucic, M, Axford, D.A, Owen, R.L, Worrall, J.A.R, Hough, M.A. | | Deposit date: | 2020-04-20 | | Release date: | 2021-01-13 | | Last modified: | 2021-01-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Serial Femtosecond Zero Dose Crystallography Captures a Water-Free Distal Heme Site in a Dye-Decolorising Peroxidase to Reveal a Catalytic Role for an Arginine in Fe IV =O Formation.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6YRJ
 
 | | SFX structure of dye-type peroxidase DtpB in the ferric state | | Descriptor: | MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE, Putative iron-dependent peroxidase | | Authors: | Lucic, M, Axford, D.A, Owen, R.L, Worrall, J.A.R, Hough, M.A. | | Deposit date: | 2020-04-20 | | Release date: | 2021-01-13 | | Last modified: | 2021-01-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Serial Femtosecond Zero Dose Crystallography Captures a Water-Free Distal Heme Site in a Dye-Decolorising Peroxidase to Reveal a Catalytic Role for an Arginine in Fe IV =O Formation.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5GVV
 
 | | Crystal structure of the glycosyltransferase GlyE in Streptococcus pneumoniae TIGR4 | | Descriptor: | Glycosyl transferase family 8, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Jiang, Y.L, Jin, H, Zhao, R.L, Yang, H.B, Chen, Y, Zhou, C.Z. | | Deposit date: | 2016-09-07 | | Release date: | 2017-03-01 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Defining the enzymatic pathway for polymorphic O-glycosylation of the pneumococcal serine-rich repeat protein PsrP.
J. Biol. Chem., 292, 2017
|
|
5GVW
 
 | | Crystal structure of the apo-form glycosyltransferase GlyE in Streptococcus pneumoniae TIGR4 | | Descriptor: | Glycosyl transferase family 8, MANGANESE (II) ION | | Authors: | Jiang, Y.L, Jin, H, Zhao, R.L, Yang, H.B, Chen, Y, Zhou, C.Z. | | Deposit date: | 2016-09-07 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Defining the enzymatic pathway for polymorphic O-glycosylation of the pneumococcal serine-rich repeat protein PsrP.
J. Biol. Chem., 292, 2017
|
|
5HQ3
 
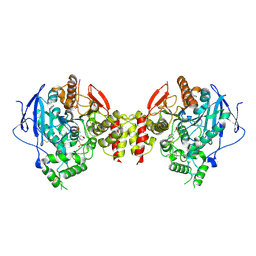 | | Stable, high-expression variant of human acetylcholinesterase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acetylcholinesterase, O-ETHYLMETHYLPHOSPHONIC ACID ESTER GROUP | | Authors: | Goldenzweig, A, Goldsmith, M, Hill, S.E, Gertman, O, Laurino, P, Ashani, Y, Dym, O, Albeck, S, Unger, T, Prilusky, J, Lieberman, R.L, Aharoni, A, Silman, I, Sussman, J.L, Tawfik, D.S, Fleishman, S.J. | | Deposit date: | 2016-01-21 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Automated Structure- and Sequence-Based Design of Proteins for High Bacterial Expression and Stability.
Mol.Cell, 63, 2016
|
|
5HGR
 
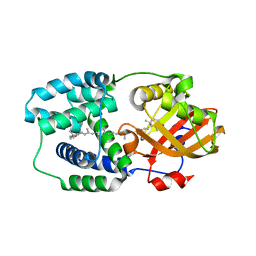 | | Structure of Anabaena (Nostoc) sp. PCC 7120 Orange Carotenoid Protein binding canthaxanthin | | Descriptor: | Orange Carotenoid Protein (OCP), beta,beta-carotene-4,4'-dione | | Authors: | Sutter, M, Leverenz, R.L, Kerfeld, C.A. | | Deposit date: | 2016-01-08 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.681 Å) | | Cite: | Different Functions of the Paralogs to the N-Terminal Domain of the Orange Carotenoid Protein in the Cyanobacterium Anabaena sp. PCC 7120.
Plant Physiol., 171, 2016
|
|
5I4S
 
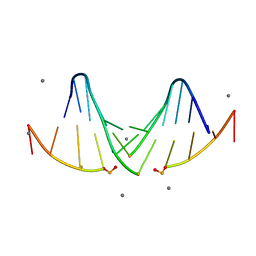 | |
5ILT
 
 | | Crystal structure of bovine Fab A01 | | Descriptor: | bovine Fab A01 heavy chain, bovine Fab A01 light chain | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2016-03-04 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conservation and diversity in the ultralong third heavy-chain complementarity-determining region of bovine antibodies.
Sci Immunol, 1, 2016
|
|
5IN9
 
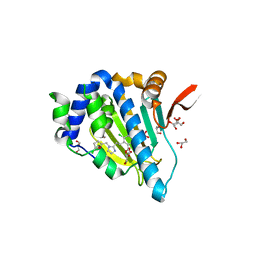 | | Crystal structure of Grp94 bound to methyl 3-chloro-2-(2-(1-((5-chlorofuran-2-yl)methyl)-1H-imidazol-2-yl)ethyl)-4,6-dihydroxybenzoate, an inhibitor based on the BnIm and Radamide scaffolds. | | Descriptor: | Endoplasmin, GLYCEROL, TRIETHYLENE GLYCOL, ... | | Authors: | Lieberman, R.L, Huard, D.J.E, Kizziah, J.L. | | Deposit date: | 2016-03-07 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Development of Glucose Regulated Protein 94-Selective Inhibitors Based on the BnIm and Radamide Scaffold.
J.Med.Chem., 59, 2016
|
|
5IJV
 
 | | Crystal structure of bovine Fab E03 | | Descriptor: | bovine Fab E03 heavy chain, bovine Fab E03 light chain | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2016-03-02 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conservation and diversity in the ultralong third heavy-chain complementarity-determining region of bovine antibodies.
Sci Immunol, 1, 2016
|
|
