2BEP
 
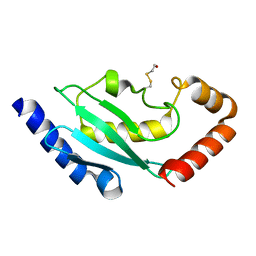 | | Crystal structure of ubiquitin conjugating enzyme E2-25K | | Descriptor: | BETA-MERCAPTOETHANOL, UBIQUITIN-CONJUGATING ENZYME E2-25 KDA | | Authors: | Pichler, A, Knipscheer, P, Oberhofer, E, Van Dijk, W.J, Korner, R, Velgaard Olsen, J, Jentsch, S, Melchior, F, Sixma, T.K. | | Deposit date: | 2004-11-29 | | Release date: | 2005-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sumo Imodification of the Ubiquitin Conjugating Enzyme E2-25K
Nat.Struct.Mol.Biol., 12, 2005
|
|
2BF8
 
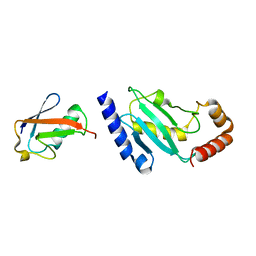 | | Crystal structure of SUMO modified ubiquitin conjugating enzyme E2- 25K | | Descriptor: | UBIQUITIN-CONJUGATING ENZYME E2-25 KDA, UBIQUITIN-LIKE PROTEIN SMT3C | | Authors: | Pichler, A, Knipscheer, P, Oberhofer, E, Van Dijk, W.J, Korner, R, Velgaard Olsen, J, Jentsch, S, Melchior, F, Sixma, T.K. | | Deposit date: | 2004-12-06 | | Release date: | 2005-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Sumo Imodification of the Ubiquitin Conjugating Enzyme E2-25K
Nat.Struct.Mol.Biol., 12, 2005
|
|
2QN0
 
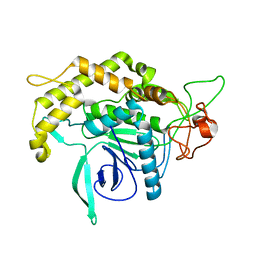 | | Structure of Botulinum neurotoxin serotype C1 light chain protease | | Descriptor: | Neurotoxin, ZINC ION | | Authors: | Jin, R, Sikorra, S, Stegmann, C.M, Pich, A, Binz, T, Brunger, A.T. | | Deposit date: | 2007-07-17 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and biochemical studies of botulinum neurotoxin serotype C1 light chain protease: implications for dual substrate specificity.
Biochemistry, 46, 2007
|
|
3R4S
 
 | | Cell entry of botulinum neurotoxin type C is dependent upon interaction with two ganglioside molecules | | Descriptor: | Botulinum neurotoxin type C1, N-acetyl-alpha-neuraminic acid, N-acetyl-beta-neuraminic acid | | Authors: | Strotmeier, J, Gu, S, Jutzi, S, Mahrhold, S, Zhou, J, Pich, A, Bigalke, H, Rummel, A, Jin, R, Binz, T. | | Deposit date: | 2011-03-17 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The biological activity of botulinum neurotoxin type C is dependent upon novel types of ganglioside binding sites.
Mol.Microbiol., 81, 2011
|
|
3R4U
 
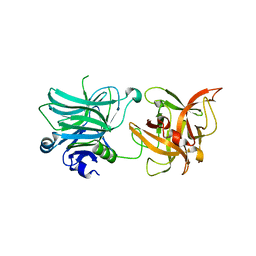 | | Cell entry of botulinum neurotoxin type C is dependent upon interaction with two ganglioside molecules | | Descriptor: | Botulinum neurotoxin type C1 | | Authors: | Strotmeier, J, Gu, S, Jutzi, S, Mahrhold, S, Zhou, J, Pich, A, Bigalke, H, Rummel, A, Jin, R, Binz, T. | | Deposit date: | 2011-03-17 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The biological activity of botulinum neurotoxin type C is dependent upon novel types of ganglioside binding sites.
Mol.Microbiol., 81, 2011
|
|
2VRR
 
 | | Structure of SUMO modified Ubc9 | | Descriptor: | FORMIC ACID, SMALL UBIQUITIN-RELATED MODIFIER 1, SODIUM ION, ... | | Authors: | Knipscheer, P, Flotho, A, Klug, H, Olsen, J.V, van Dijk, W.J, Fish, A, Johnson, E.S, Mann, M, Sixma, T.K, Pichler, A. | | Deposit date: | 2008-04-13 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Ubc9 sumoylation regulates SUMO target discrimination.
Mol. Cell, 31, 2008
|
|
4HOR
 
 | | Crystal Structure of Full-Length Human IFIT5 with 5`-triphosphate Oligocytidine | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 5, MAGNESIUM ION, RNA (5'-R(*(CTP)P*CP*CP*CP*C)-3') | | Authors: | Abbas, Y.M, Pichlmair, A, Gorna, M.W, Superti-Furga, G, Nagar, B. | | Deposit date: | 2012-10-22 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.861 Å) | | Cite: | Structural basis for viral 5'-PPP-RNA recognition by human IFIT proteins.
Nature, 494, 2013
|
|
4HOU
 
 | | Crystal Structure of N-terminal Human IFIT1 | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 1 | | Authors: | Abbas, Y.M, Pichlmair, A, Gorna, M.W, Superti-Furga, G, Nagar, B. | | Deposit date: | 2012-10-22 | | Release date: | 2013-01-30 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for viral 5'-PPP-RNA recognition by human IFIT proteins.
Nature, 494, 2013
|
|
4HOT
 
 | | Crystal Structure of Full-Length Human IFIT5 with 5`-triphosphate Oligoadenine | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 5, MAGNESIUM ION, RNA (5'-R(*(ATP)P*AP*AP*A)-3') | | Authors: | Abbas, Y.M, Pichlmair, A, Gorna, M.W, Superti-Furga, G, Nagar, B. | | Deposit date: | 2012-10-22 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural basis for viral 5'-PPP-RNA recognition by human IFIT proteins.
Nature, 494, 2013
|
|
4HOS
 
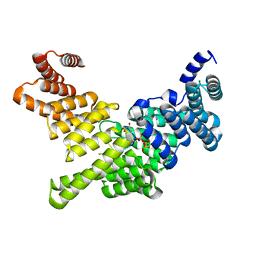 | | Crystal Structure of Full-Length Human IFIT5 with 5`-triphosphate Oligouridine | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 5, RNA (5'-R(*(UTP)P*UP*UP*U)-3'), SODIUM ION | | Authors: | Abbas, Y.M, Pichlmair, A, Gorna, M.W, Superti-Furga, G, Nagar, B. | | Deposit date: | 2012-10-22 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for viral 5'-PPP-RNA recognition by human IFIT proteins.
Nature, 494, 2013
|
|
4HOQ
 
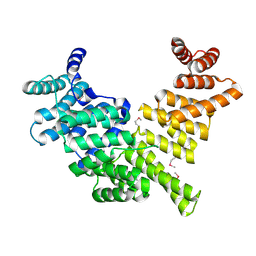 | | Crystal Structure of Full-Length Human IFIT5 | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 5 | | Authors: | Abbas, Y.M, Pichlmair, A, Gorna, M.W, Superti-Furga, G, Nagar, B. | | Deposit date: | 2012-10-22 | | Release date: | 2013-01-23 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis for viral 5'-PPP-RNA recognition by human IFIT proteins.
Nature, 494, 2013
|
|
3E3U
 
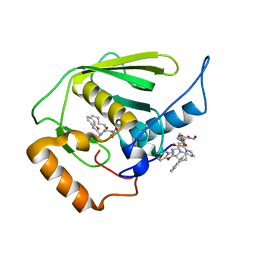 | | Crystal structure of Mycobacterium tuberculosis peptide deformylase in complex with inhibitor | | Descriptor: | N-[(2R)-2-{[(2S)-2-(1,3-benzoxazol-2-yl)pyrrolidin-1-yl]carbonyl}hexyl]-N-hydroxyformamide, NICKEL (II) ION, Peptide deformylase | | Authors: | Meng, W, Xu, M, Pan, S, Koehn, J. | | Deposit date: | 2008-08-08 | | Release date: | 2009-01-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Peptide deformylase inhibitors of Mycobacterium tuberculosis: synthesis, structural investigations, and biological results.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
8B4A
 
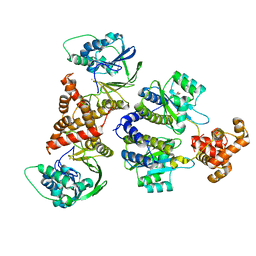 | | Nativ complex of PqsE and RhlR with autoinducer C4-HSL | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, FE (III) ION, N-[(3S)-2-oxotetrahydrofuran-3-yl]butanamide, ... | | Authors: | Borgert, S.R, Blankenfeldt, W. | | Deposit date: | 2022-09-20 | | Release date: | 2022-12-14 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
3OBR
 
 | |
3OBT
 
 | |
7R3H
 
 | |
7R3J
 
 | | Nativ complex of PqsE and RhlR with the synthetic antagonist mBTL | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide, FE (III) ION, ... | | Authors: | Borgert, S.R, Schmelz, S, Blankenfeldt, W. | | Deposit date: | 2022-02-07 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
7R3E
 
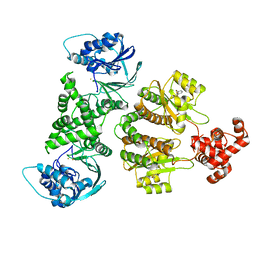 | | Fusion construct of PqsE and RhlR in complex with the synthetic antagonist mBTL | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase,Regulatory protein RhlR, 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide, FE (III) ION | | Authors: | Borgert, S.R, Schmelz, S, Blankenfeldt, W. | | Deposit date: | 2022-02-07 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
7R3F
 
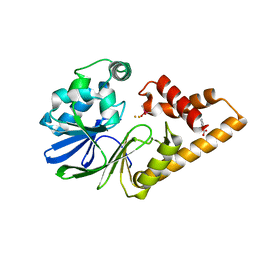 | | Monomeric PqsE mutant E187R | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, BENZOIC ACID, CACODYLATE ION, ... | | Authors: | Borgert, S.R, Schmelz, S, Blankenfeldt, W. | | Deposit date: | 2022-02-07 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
7R3G
 
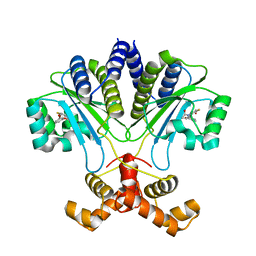 | |
7R3I
 
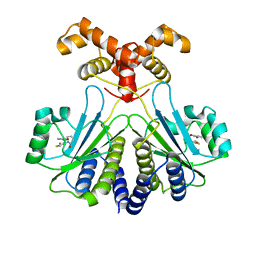 | |
5D2M
 
 | | Complex between human SUMO2-RANGAP1, UBC9 and ZNF451 | | Descriptor: | 1,2-ETHANEDIOL, Ran GTPase-activating protein 1, SUMO-conjugating enzyme UBC9, ... | | Authors: | Cappadocia, L, Lima, C.D. | | Deposit date: | 2015-08-05 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for catalytic activation by the human ZNF451 SUMO E3 ligase.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6HVH
 
 | | Human PFKFB3 in complex with a N-Aryl 6-Aminoquinoxaline inhibitor 1 | | Descriptor: | 3-[[8-(1-methylindol-6-yl)quinoxalin-6-yl]amino]-~{N}-(oxan-4-yl)pyridine-4-carboxamide, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, ... | | Authors: | Banaszak, K, Jakubiec, K, Bialas, A, Fabritius, C.H, Nowak, M. | | Deposit date: | 2018-10-11 | | Release date: | 2018-11-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Discovery and Structure-Activity Relationships of N-Aryl 6-Aminoquinoxalines as Potent PFKFB3 Kinase Inhibitors.
ChemMedChem, 14, 2019
|
|
6IBZ
 
 | | Human PFKFB3 in complex with a N-Aryl 6-Aminoquinoxaline inhibitor 7 | | Descriptor: | 2-[[8-(1-methylindol-6-yl)quinoxalin-6-yl]amino]-~{N}-(pyrimidin-5-ylmethyl)benzenesulfonamide, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, ... | | Authors: | Banaszak, K, Tomczyk, M, Guzik, P, Fabritius, C.H, Nowak, M. | | Deposit date: | 2018-12-01 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Synthesis of amide and sulfonamide substituted N-aryl 6-aminoquinoxalines as PFKFB3 inhibitors with improved physicochemical properties.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
6IBY
 
 | | Human PFKFB3 in complex with a N-Aryl 6-Aminoquinoxaline inhibitor 6 | | Descriptor: | 3-[[8-(1-methylindol-6-yl)quinoxalin-6-yl]amino]-~{N}-[(3~{S})-1-methylpyrrolidin-3-yl]pyridine-4-carboxamide, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, ... | | Authors: | Banaszak, K, Pawlik, H, Bialas, A, Fabritius, C.H, Nowak, M. | | Deposit date: | 2018-12-01 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Synthesis of amide and sulfonamide substituted N-aryl 6-aminoquinoxalines as PFKFB3 inhibitors with improved physicochemical properties.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
