1WUF
 
 | | Crystal structure of protein GI:16801725, member of Enolase superfamily from Listeria innocua Clip11262 | | Descriptor: | MAGNESIUM ION, hypothetical protein lin2664 | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-07 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2FH7
 
 | | Crystal structure of the phosphatase domains of human PTP SIGMA | | Descriptor: | Receptor-type tyrosine-protein phosphatase S | | Authors: | Alvarado, J, Udupi, R, Smith, D, Koss, J, Wasserman, S.R, Ozyurt, S, Atwell, S, Powell, A, Kearins, M.C, Rooney, I, Maletic, M, Bain, K.T, Freeman, J.C, Russell, M, Thompson, D.A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-12-23 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2G59
 
 | |
2HHL
 
 | | Crystal structure of the human small CTD phosphatase 3 isoform 1 | | Descriptor: | 12-TUNGSTOPHOSPHATE, CTD small phosphatase-like protein | | Authors: | Malashkevich, V.N, Toro, R, Ramagopal, U, Sauder, J.M, Schwinn, K.D, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-06-28 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2HY3
 
 | | Crystal structure of the human tyrosine receptor phosphate gamma in complex with vanadate | | Descriptor: | Receptor-type tyrosine-protein phosphatase gamma, VANADATE ION | | Authors: | Jin, X, Min, T, Bera, A, Mu, H, Sauder, J.M, Freeman, J.C, Reyes, C, Smith, D, Wasserman, S.R, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-04 | | Release date: | 2006-09-05 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2I44
 
 | |
2HXP
 
 | | Crystal Structure of the human phosphatase (DUSP9) | | Descriptor: | Dual specificity protein phosphatase 9, PHOSPHATE ION | | Authors: | Madegowda, M, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-03 | | Release date: | 2006-08-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
2I0O
 
 | | Crystal structure of Anopheles gambiae Ser/Thr phosphatase complexed with Zn2+ | | Descriptor: | Ser/Thr phosphatase, ZINC ION | | Authors: | Jin, X, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-10 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2IRM
 
 | | Crystal structure of mitogen-activated protein kinase kinase kinase 7 interacting protein 1 from Anopheles gambiae | | Descriptor: | mitogen-activated protein kinase kinase kinase 7 interacting protein 1 | | Authors: | Jin, X, Bonanno, J.B, Pelletier, L, Freeman, J.C, Wasserman, S, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-15 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2ISN
 
 | | Crystal structure of a phosphatase from a pathogenic strain Toxoplasma gondii | | Descriptor: | NYSGXRC-8828z, phosphatase, PRASEODYMIUM ION, ... | | Authors: | Agarwal, R, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-18 | | Release date: | 2006-10-31 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2IQ1
 
 | | Crystal structure of human PPM1K | | Descriptor: | MAGNESIUM ION, Protein phosphatase 2C kappa, PPM1K | | Authors: | Bonanno, J.B, Freeman, J, Russell, M, Bain, K.T, Adams, J, Pelletier, L, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-12 | | Release date: | 2006-11-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural genomics of protein phosphatases
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
4EJ6
 
 | |
4EJM
 
 | |
4F62
 
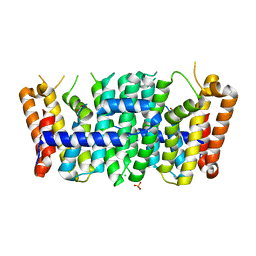 | | Crystal structure of a putative farnesyl-diphosphate synthase from Marinomonas sp. MED121 (Target EFI-501980) | | Descriptor: | CHLORIDE ION, GLYCEROL, Geranyltranstransferase, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4GEK
 
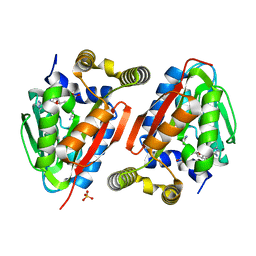 | | Crystal Structure of wild-type CmoA from E.coli | | Descriptor: | (2S)-4-[{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}(carboxylatomethyl)sulfonio] -2-ammoniobutanoate, SULFATE ION, tRNA (cmo5U34)-methyltransferase | | Authors: | Kim, J, Toro, R, Bonanno, J.B, Bhosle, R, Sampathkumar, P, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-08-02 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided discovery of the metabolite carboxy-SAM that modulates tRNA function
Nature, 498, 2013
|
|
2NV5
 
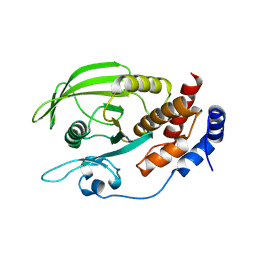 | | Crystal structure of a C-terminal phosphatase domain of Rattus norvegicus ortholog of human protein tyrosine phosphatase, receptor type, D (PTPRD) | | Descriptor: | PTPRD, PHOSPHATASE | | Authors: | Bonanno, J.B, Gilmore, J, Bain, K.T, Iizuka, M, Xu, W, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-10 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
4HGP
 
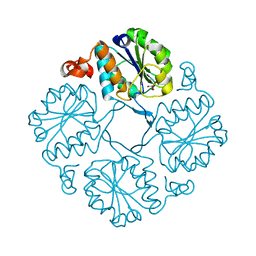 | |
4HGQ
 
 | |
4HGO
 
 | |
4HGN
 
 | |
4HGR
 
 | |
2OYC
 
 | | Crystal structure of human pyridoxal phosphate phosphatase | | Descriptor: | Pyridoxal phosphate phosphatase, SODIUM ION, TUNGSTATE(VI)ION | | Authors: | Ramagopal, U.A, Freeman, J, Izuka, M, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-21 | | Release date: | 2007-03-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
2P69
 
 | | Crystal Structure of Human Pyridoxal Phosphate Phosphatase with PLP | | Descriptor: | CALCIUM ION, PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate phosphatase | | Authors: | Ramagopal, U.A, Freeman, J, Izuka, M, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
2P8E
 
 | | Crystal structure of the serine/threonine phosphatase domain of human PPM1B | | Descriptor: | MAGNESIUM ION, PPM1B beta isoform variant 6 | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Lau, C, Xu, W, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-22 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
2OZT
 
 | | Crystal structure of O-succinylbenzoate synthase from Thermosynechococcus elongatus BP-1 | | Descriptor: | PHOSPHATE ION, SODIUM ION, Tlr1174 protein | | Authors: | Malashkevich, V.N, Bonanno, J, Toro, R, Sauder, J.M, Schwinn, K.D, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Gheyi, T, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-27 | | Release date: | 2007-03-13 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
