1Z0M
 
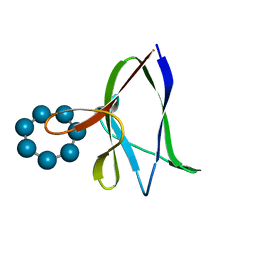 | | the glycogen-binding domain of the AMP-activated protein kinase beta1 subunit | | Descriptor: | 5'-AMP-activated protein kinase, beta-1 subunit, Cycloheptakis-(1-4)-(alpha-D-glucopyranose) | | Authors: | Polekhina, G, Gupta, A, van Denderen, B.J, Feil, S.C, Kemp, B.E, Stapleton, D, Parker, M.W. | | Deposit date: | 2005-03-02 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural Basis for Glycogen Recognition by AMP-Activated Protein Kinase.
Structure, 13, 2005
|
|
1Z0N
 
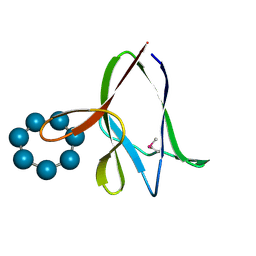 | | the glycogen-binding domain of the AMP-activated protein kinase | | Descriptor: | 5'-AMP-activated protein kinase, beta-1 subunit, Cycloheptakis-(1-4)-(alpha-D-glucopyranose) | | Authors: | Polekhina, G, Gupta, A, van Denderen, B.J, Feil, S.C, Kemp, B.E, Stapleton, D, Parker, M.W. | | Deposit date: | 2005-03-02 | | Release date: | 2005-10-25 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural Basis for Glycogen Recognition by AMP-Activated Protein Kinase.
Structure, 13, 2005
|
|
20GS
 
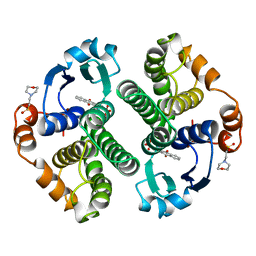 | | GLUTATHIONE S-TRANSFERASE P1-1 COMPLEXED WITH CIBACRON BLUE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CIBACRON BLUE, GLUTATHIONE S-TRANSFERASE | | Authors: | Oakley, A.J, Lo Bello, M, Nuccetelli, M, Mazzetti, A.P, Parker, M.W. | | Deposit date: | 1997-12-16 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The ligandin (non-substrate) binding site of human Pi class glutathione transferase is located in the electrophile binding site (H-site).
J.Mol.Biol., 291, 1999
|
|
7RQ0
 
 | |
5IMT
 
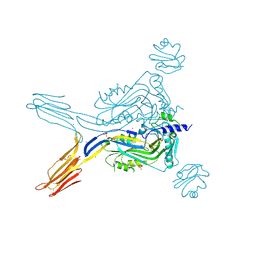 | | Toxin receptor complex | | Descriptor: | CD59 glycoprotein, COPPER (II) ION, Intermedilysin, ... | | Authors: | Morton, C.J, Lawrence, S.L, Feil, S.C, Parker, M.W. | | Deposit date: | 2016-03-06 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7001 Å) | | Cite: | Structural Basis for Receptor Recognition by the Human CD59-Responsive Cholesterol-Dependent Cytolysins.
Structure, 24, 2016
|
|
5IMW
 
 | | Trapped Toxin | | Descriptor: | Intermedilysin | | Authors: | Lawrence, S.L, Feil, S.C, Morton, C.J, Parker, M.W. | | Deposit date: | 2016-03-07 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural Basis for Receptor Recognition by the Human CD59-Responsive Cholesterol-Dependent Cytolysins.
Structure, 24, 2016
|
|
2FK3
 
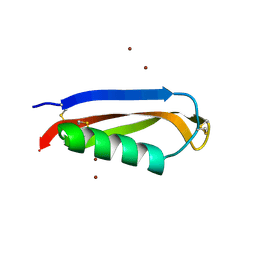 | |
2FJZ
 
 | |
2FKL
 
 | |
5IMY
 
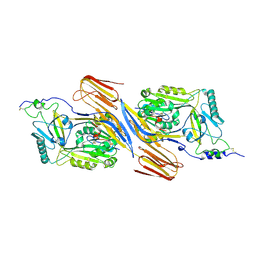 | | Trapped Toxin | | Descriptor: | CD59 glycoprotein, Vaginolysin | | Authors: | Lawrence, S.L, Morton, C.J, Parker, M.W. | | Deposit date: | 2016-03-07 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Receptor Recognition by the Human CD59-Responsive Cholesterol-Dependent Cytolysins.
Structure, 24, 2016
|
|
2FK1
 
 | |
2FK2
 
 | |
2FHE
 
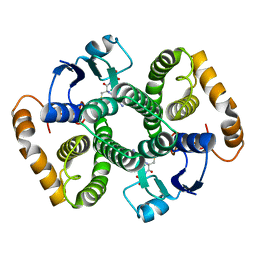 | | FASCIOLA HEPATICA GLUTATHIONE S-TRANSFERASE ISOFORM 1 IN COMPLEX WITH GLUTATHIONE | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE | | Authors: | Polekhina, G, Rossjohn, J, Feil, S.C, Parker, M.W. | | Deposit date: | 1998-10-21 | | Release date: | 1998-10-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallization, structural determination and analysis of a novel parasite vaccine candidate: Fasciola hepatica glutathione S-transferase.
J.Mol.Biol., 273, 1997
|
|
2HGS
 
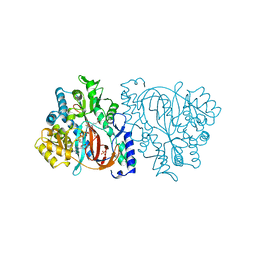 | | HUMAN GLUTATHIONE SYNTHETASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLUTATHIONE, MAGNESIUM ION, ... | | Authors: | Polekhina, G, Board, P, Rossjohn, J, Parker, M.W. | | Deposit date: | 1999-01-04 | | Release date: | 1999-06-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis of glutathione synthetase deficiency and a rare gene permutation event.
EMBO J., 18, 1999
|
|
1FHE
 
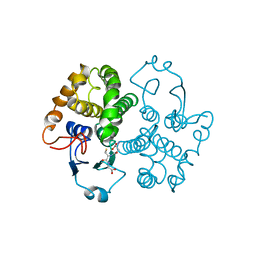 | | GLUTATHIONE TRANSFERASE (FH47) FROM FASCIOLA HEPATICA | | Descriptor: | GLUTATHIONE, GLUTATHIONE TRANSFERASE | | Authors: | Rossjohn, J, Parker, M.W. | | Deposit date: | 1997-07-24 | | Release date: | 1998-07-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallization, structural determination and analysis of a novel parasite vaccine candidate: Fasciola hepatica glutathione S-transferase.
J.Mol.Biol., 273, 1997
|
|
1GSS
 
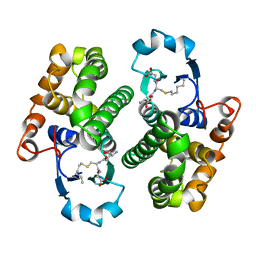 | | THREE-DIMENSIONAL STRUCTURE OF CLASS PI GLUTATHIONE S-TRANSFERASE FROM HUMAN PLACENTA IN COMPLEX WITH S-HEXYLGLUTATHIONE AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | GLUTATHIONE S-TRANSFERASE, L-gamma-glutamyl-S-hexyl-L-cysteinylglycine | | Authors: | Reinemer, P, Dirr, H.W, Ladenstein, R, Lobello, M, Federici, G, Huber, R, Parker, M.W. | | Deposit date: | 1992-05-28 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of class pi glutathione S-transferase from human placenta in complex with S-hexylglutathione at 2.8 A resolution.
J.Mol.Biol., 227, 1992
|
|
1K2F
 
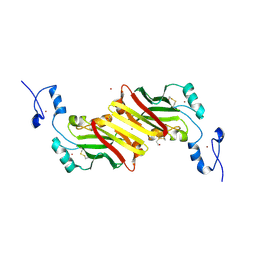 | | siah, Seven In Absentia Homolog | | Descriptor: | BETA-MERCAPTOETHANOL, ZINC ION, siah-1A protein | | Authors: | Polekhina, G, House, C.M, Traficante, N, Mackay, J.P, Relaix, F, Sassoon, D.A, Parker, M.W, Bowtell, D.D.L. | | Deposit date: | 2001-09-26 | | Release date: | 2001-12-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Siah ubiquitin ligase is structurally related to TRAF and modulates TNF-alpha signaling.
Nat.Struct.Biol., 9, 2002
|
|
1KBN
 
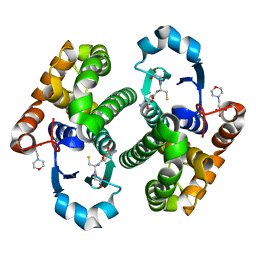 | | Glutathione transferase mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, GLYCEROL, ... | | Authors: | Rossjohn, J, Parker, M.W. | | Deposit date: | 2001-11-06 | | Release date: | 2003-11-11 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Glutathione transferase mutant
To be Published
|
|
3GSS
 
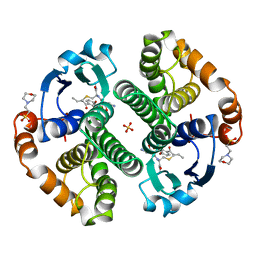 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1 IN COMPLEX WITH ETHACRYNIC ACID-GLUTATHIONE CONJUGATE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ETHACRYNIC ACID, GLUTATHIONE, ... | | Authors: | Oakley, A.J, Rossjohn, J, Parker, M.W. | | Deposit date: | 1996-10-29 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The three-dimensional structure of the human Pi class glutathione transferase P1-1 in complex with the inhibitor ethacrynic acid and its glutathione conjugate.
Biochemistry, 36, 1997
|
|
4OJF
 
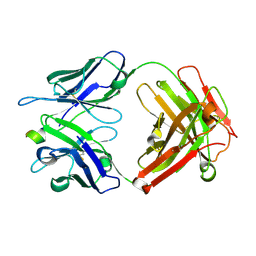 | | Humanised 3D6 Fab complexed to amyloid beta 1-8 | | Descriptor: | Amyloid beta A4 protein, Humanised 3D6 Fab Heavy Chain, Humanised 3D6 Fab Light Chain | | Authors: | Miles, L.A, Crespi, G.A.N, Parker, M.W. | | Deposit date: | 2014-01-21 | | Release date: | 2015-01-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystallization and preliminary X-ray diffraction analysis of the Fab portion of the Alzheimer's disease immunotherapy candidate bapineuzumab complexed with amyloid-beta
ACTA CRYSTALLOGR.,SECT.F, 70, 2014
|
|
19GS
 
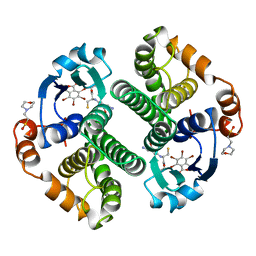 | | Glutathione s-transferase p1-1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,3'-(4,5,6,7-TETRABROMO-3-OXO-1(3H)-ISOBENZOFURANYLIDENE)BIS [6-HYDROXYBENZENESULFONIC ACID]ANION, GLUTATHIONE, ... | | Authors: | Oakley, A.J, Lo Bello, M, Parker, M.W. | | Deposit date: | 1997-12-14 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The ligandin (non-substrate) binding site of human Pi class glutathione transferase is located in the electrophile binding site (H-site).
J.Mol.Biol., 291, 1999
|
|
1A87
 
 | | COLICIN N | | Descriptor: | COLICIN N | | Authors: | Vetter, I.R, Parker, M.W, Tucker, A.D, Lakey, J.H, Pattus, F, Tsernoglou, D. | | Deposit date: | 1998-04-03 | | Release date: | 1999-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of a colicin N fragment suggests a model for toxicity.
Structure, 6, 1998
|
|
2LJR
 
 | | GLUTATHIONE TRANSFERASE APO-FORM FROM HUMAN | | Descriptor: | GLUTATHIONE S-TRANSFERASE | | Authors: | Rossjohn, J, Mckinstry, W.J, Oakley, A.J, Verger, D, Flanagan, J, Chelvanayagam, G, Tan, K.L, Board, P.G, Parker, M.W. | | Deposit date: | 1998-03-08 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Human theta class glutathione transferase: the crystal structure reveals a sulfate-binding pocket within a buried active site.
Structure, 6, 1998
|
|
2PMT
 
 | | GLUTATHIONE TRANSFERASE FROM PROTEUS MIRABILIS | | Descriptor: | GLUTATHIONE, GLUTATHIONE TRANSFERASE | | Authors: | Rossjohn, J, Polekhina, G, Feil, S.C, Allocati, N, Masulli, M, Diilio, C, Parker, M.W. | | Deposit date: | 1998-04-28 | | Release date: | 1999-04-27 | | Last modified: | 2012-01-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A mixed disulfide bond in bacterial glutathione transferase: functional and evolutionary implications.
Structure, 6, 1998
|
|
1KOA
 
 | | TWITCHIN KINASE FRAGMENT (C.ELEGANS), AUTOREGULATED PROTEIN KINASE AND IMMUNOGLOBULIN DOMAINS | | Descriptor: | TWITCHIN | | Authors: | Kobe, B, Heierhorst, J, Feil, S.C, Parker, M.W, Benian, G.M, Weiss, K.R, Kemp, B.E. | | Deposit date: | 1996-06-28 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Giant protein kinases: domain interactions and structural basis of autoregulation.
EMBO J., 15, 1996
|
|
