4R27
 
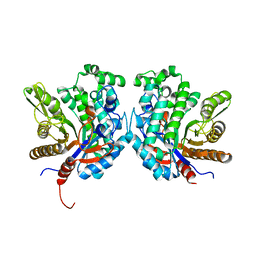 | | Crystal structure of beta-glycosidase BGL167 | | Descriptor: | Glycoside hydrolase | | Authors: | Park, S.J, Choi, J.M, Kyeong, H.H, Kim, S.G, Kim, H.S. | | Deposit date: | 2014-08-09 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Rational design of a beta-glycosidase with high regiospecificity for triterpenoid tailoring
Chembiochem, 16, 2015
|
|
2H9Z
 
 | | Solution structure of hypothetical protein, HP0495 from Helicobacter pylori | | Descriptor: | Hypothetical protein HP0495 | | Authors: | Seo, M.D, Park, S.J, Kim, H.J, Lee, B.J. | | Deposit date: | 2006-06-12 | | Release date: | 2007-05-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hypothetical protein, HP0495 (Y495_HELPY) from Helicobacter pylori.
Proteins, 67, 2007
|
|
1ZHC
 
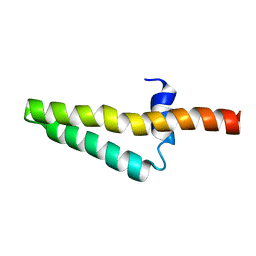 | |
1YG0
 
 | |
1Z8M
 
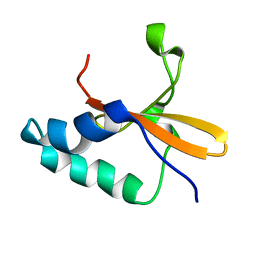 | |
1Q3O
 
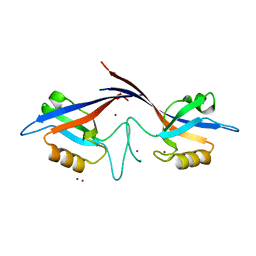 | | Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization | | Descriptor: | BROMIDE ION, Shank1 | | Authors: | Im, Y.J, Lee, J.H, Park, S.H, Park, S.J, Rho, S.-H, Kang, G.B, Kim, E, Eom, S.H. | | Deposit date: | 2003-07-31 | | Release date: | 2004-01-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization
J.Biol.Chem., 278, 2003
|
|
1Q3P
 
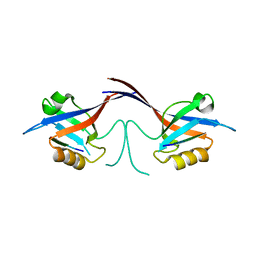 | | Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization | | Descriptor: | C-terminal hexapeptide from Guanylate kinase-associated protein, Shank1 | | Authors: | Im, Y.J, Lee, J.H, Park, S.H, Park, S.J, Rho, S.-H, Kang, G.B, Kim, E, Eom, S.H. | | Deposit date: | 2003-07-31 | | Release date: | 2004-01-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization
J.Biol.Chem., 278, 2003
|
|
3QJM
 
 | | Structural flexibility of Shank PDZ domain is important for its binding to different ligands | | Descriptor: | Beta-PIX, SH3 and multiple ankyrin repeat domains protein 1 | | Authors: | Lee, J.H, Park, H, Park, S.J, Kim, H.J, Eom, S.H. | | Deposit date: | 2011-01-30 | | Release date: | 2011-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | The structural flexibility of the shank1 PDZ domain is important for its binding to different ligands
Biochem.Biophys.Res.Commun., 407, 2011
|
|
3QJN
 
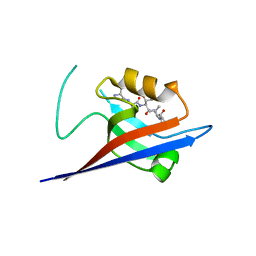 | | Structural flexibility of Shank PDZ domain is important for its binding to different ligands | | Descriptor: | Beta-PIX, SH3 and multiple ankyrin repeat domains protein 1 | | Authors: | Lee, J.H, Park, H, Park, S.J, Kim, H.J, Eom, S.H. | | Deposit date: | 2011-01-30 | | Release date: | 2011-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The structural flexibility of the shank1 PDZ domain is important for its binding to different ligands
Biochem.Biophys.Res.Commun., 407, 2011
|
|
2OTR
 
 | |
2R62
 
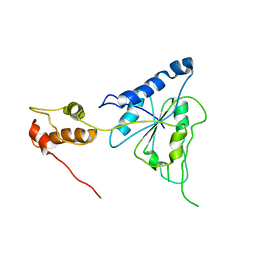 | | Crystal structure of Helicobacter pylori ATP dependent protease, FtsH | | Descriptor: | Cell division protease ftsH homolog | | Authors: | Kim, S.H, Kang, G.B, Song, H.-E, Park, S.J, Bae, M.-H, Eom, S.H. | | Deposit date: | 2007-09-05 | | Release date: | 2008-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural studies on Helicobacter pyloriATP-dependent protease, FtsH
J.SYNCHROTRON RADIAT., 15, 2008
|
|
2R65
 
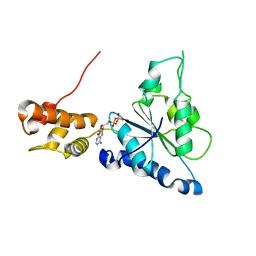 | | Crystal structure of Helicobacter pylori ATP dependent protease, FtsH ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division protease ftsH homolog | | Authors: | Kim, S.H, Kang, G.B, Song, H.-E, Park, S.J, Bae, M.-H, Eom, S.H. | | Deposit date: | 2007-09-05 | | Release date: | 2008-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural studies on Helicobacter pyloriATP-dependent protease, FtsH
J.SYNCHROTRON RADIAT., 15, 2008
|
|
6IFM
 
 | | Crystal structure of DNA bound VapBC from Salmonella typhimurium | | Descriptor: | Antitoxin VapB, DNA backward (27-MER), DNA forward (27-MER), ... | | Authors: | Park, D.W, Lee, B.J. | | Deposit date: | 2018-09-20 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Crystal structure of proteolyzed VapBC and DNA-bound VapBC from Salmonella enterica Typhimurium LT2 and VapC as a putative Ca2+-dependent ribonuclease.
Faseb J., 34, 2020
|
|
6IFC
 
 | | Crystal structure of VapBC from Salmonella typhimurium | | Descriptor: | Antitoxin VapB, CALCIUM ION, tRNA(fMet)-specific endonuclease VapC | | Authors: | Park, D.W, Lee, B.J. | | Deposit date: | 2018-09-19 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of proteolyzed VapBC and DNA-bound VapBC from Salmonella enterica Typhimurium LT2 and VapC as a putative Ca2+-dependent ribonuclease.
Faseb J., 34, 2020
|
|
6IM5
 
 | | YAP-binding domain of human TEAD1 | | Descriptor: | PHOSPHATE ION, Transcriptional enhancer factor TEF-1 | | Authors: | Mo, Y, Lee, H.S, Kim, S.J, Ku, B. | | Deposit date: | 2018-10-22 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Crystal Structure of the YAP-binding Domain of Human TEAD1
Bull.Korean Chem.Soc., 40, 2019
|
|
3UI3
 
 | |
9JUB
 
 | | Cryo-EM structure of human hyaluronidase PH-20 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 79C11Fab, ... | | Authors: | Im, S.-B, Jeong, T.-K, Kim, N, Oh, B.-H. | | Deposit date: | 2024-10-07 | | Release date: | 2024-10-30 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM Structure of Human Hyaluronidase PH-20.
Proteins, 93, 2025
|
|
7CM4
 
 | | Crystal Structure of COVID-19 virus spike receptor-binding domain complexed with a neutralizing antibody CT-P59 | | Descriptor: | 1,2-ETHANEDIOL, IgG heavy chain, IgG light chain, ... | | Authors: | Kim, Y.G, Jeong, J.H, Bae, J.S, Lee, J. | | Deposit date: | 2020-07-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein.
Nat Commun, 12, 2021
|
|
5Y7X
 
 | | Human Peroxisome proliferator-activated receptor (PPAR) delta in complexed with a potent and selective agonist | | Descriptor: | 2-[2-methyl-4-[[4-methyl-2-[4-(trifluoromethyl)phenyl]-1,3-selenazol-5-yl]methylsulfanyl]phenoxy]ethanoic acid, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Kim, H.L, Chin, J.W, Cho, S.J, Song, J.Y, Yoon, H.S, Bae, J.H. | | Deposit date: | 2017-08-18 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Highly potent and selective PPAR delta agonist reverses memory deficits in mouse models of Alzheimer's disease.
Theranostics, 14, 2024
|
|
5XE2
 
 | | Endoribonuclease from Mycobacterial species | | Descriptor: | Endoribonuclease MazF4 | | Authors: | Ahn, D.-H, Lee, K.-Y, Lee, S.J, Yoon, H.J, Kim, S.-J, Lee, B.-J. | | Deposit date: | 2017-03-31 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural analyses of the MazEF4 toxin-antitoxin pair in Mycobacterium tuberculosis provide evidence for a unique extracellular death factor.
J. Biol. Chem., 292, 2017
|
|
5XE3
 
 | | Endoribonuclease in complex with its cognate antitoxin from Mycobacterial species | | Descriptor: | Endoribonuclease MazF4, Probable antitoxin MazE4 | | Authors: | Ahn, D.-H, Lee, K.-Y, Lee, S.J, Yoon, H.J, Kim, S.-J, Lee, B.-J. | | Deposit date: | 2017-03-31 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analyses of the MazEF4 toxin-antitoxin pair in Mycobacterium tuberculosis provide evidence for a unique extracellular death factor.
J. Biol. Chem., 292, 2017
|
|
6JKG
 
 | | The NAD+-free form of human NSDHL | | Descriptor: | Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating | | Authors: | Kim, D, Lee, S.J, Lee, B. | | Deposit date: | 2019-02-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of human NSDHL and development of its novel inhibitor with the potential to suppress EGFR activity.
Cell.Mol.Life Sci., 78, 2021
|
|
6JKH
 
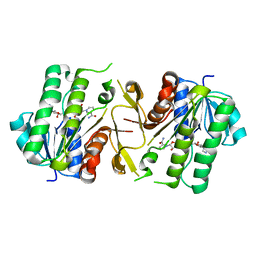 | | The NAD+-bound form of human NSDHL | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating | | Authors: | Kim, D, Lee, S.J, Lee, B. | | Deposit date: | 2019-02-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of human NSDHL and development of its novel inhibitor with the potential to suppress EGFR activity.
Cell.Mol.Life Sci., 78, 2021
|
|
9B94
 
 | |
9B91
 
 | |
