1U2N
 
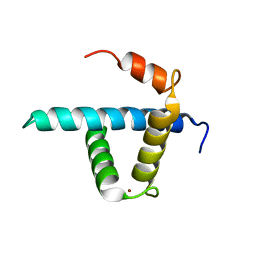 | | Structure CBP TAZ1 Domain | | Descriptor: | CREB binding protein, ZINC ION | | Authors: | De Guzman, R.N, Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2004-07-19 | | Release date: | 2005-04-26 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | CBP/p300 TAZ1 domain forms a structured scaffold for ligand binding
Biochemistry, 44, 2005
|
|
1TOT
 
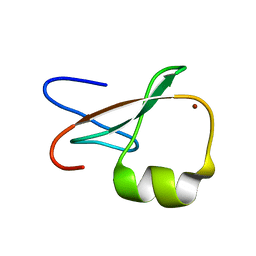 | | ZZ Domain of CBP- a Novel Fold for a Protein Interaction Module | | Descriptor: | CREB-binding protein, ZINC ION | | Authors: | Legge, G.B, Martinez-Yamout, M.A, Hambly, D.M, Trinh, T, Dyson, H.J, Wright, P.E. | | Deposit date: | 2004-06-15 | | Release date: | 2005-01-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | ZZ domain of CBP: an unusual zinc finger fold in a protein interaction module
J.Mol.Biol., 343, 2004
|
|
1UUB
 
 | |
1UUA
 
 | |
1KD6
 
 | | Solution structure of the eukaryotic pore-forming cytolysin equinatoxin II | | Descriptor: | EQUINATOXIN II | | Authors: | Hinds, M.G, Zhang, W, Anderluh, G, Hansen, P.E, Norton, R.S. | | Deposit date: | 2001-11-12 | | Release date: | 2002-02-13 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the eukaryotic pore-forming cytolysin equinatoxin II: implications for pore formation.
J.Mol.Biol., 315, 2002
|
|
1MYF
 
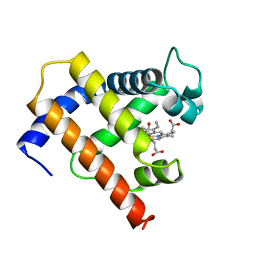 | | SOLUTION STRUCTURE OF CARBONMONOXY MYOGLOBIN DETERMINED FROM NMR DISTANCE AND CHEMICAL SHIFT CONSTRAINTS | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Osapay, K, Theriault, Y, Wright, P.E, Case, D.A. | | Deposit date: | 1994-12-02 | | Release date: | 1995-02-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of carbonmonoxy myoglobin determined from nuclear magnetic resonance distance and chemical shift constraints.
J.Mol.Biol., 244, 1994
|
|
1OVA
 
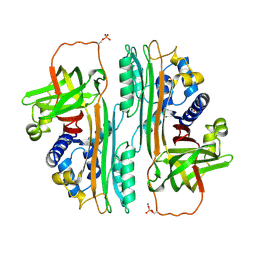 | |
6T1M
 
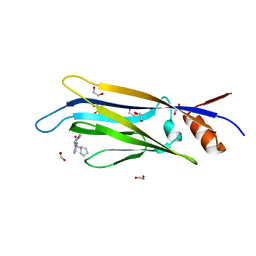 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 4 | | Descriptor: | 1,2-ETHANEDIOL, 4-cyano-~{N}-[2-(piperidin-1-ylmethyl)-1~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
6T1O
 
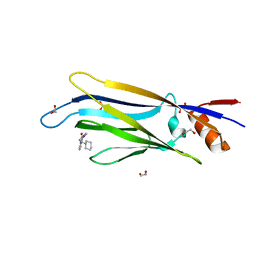 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 6 | | Descriptor: | 1,2-ETHANEDIOL, 4-iodanyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
6T1N
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 5 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
6UNA
 
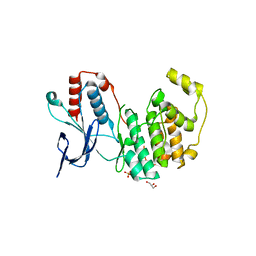 | | Crystal structure of inactive p38gamma | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 12, SULFATE ION | | Authors: | Aoto, P.C, Stanfield, R.L, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2019-10-11 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | A Dynamic Switch in Inactive p38 gamma Leads to an Excited State on the Pathway to an Active Kinase.
Biochemistry, 58, 2019
|
|
6UXO
 
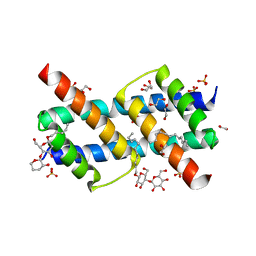 | | Crystal structure of BAK core domain BH3-groove-dimer in complex with DDM | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Bcl-2 homologous antagonist/killer, ... | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-11-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | BAK core dimers bind lipids and can be bridged by them.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6V8U
 
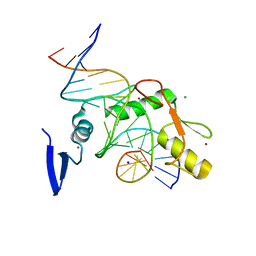 | | Kaiso (ZBTB33) zinc finger DNA binding domain in complex with a modified Kaiso binding sequence (KBS) | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*GP*GP*CP*AP*CP*GP*AP*AP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*TP*CP*GP*TP*GP*CP*CP*AP*AP*TP*AP*AP*CP*G)-3'), ... | | Authors: | Nikolova, E.N, Stanfield, R.L, Dyson, H.J, Wright, P.E. | | Deposit date: | 2019-12-12 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | A Conformational Switch in the Zinc Finger Protein Kaiso Mediates Differential Readout of Specific and Methylated DNA Sequences.
Biochemistry, 59, 2020
|
|
6UXQ
 
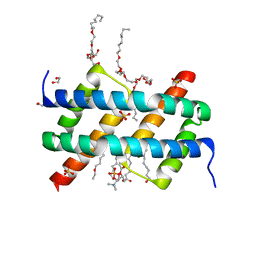 | | Crystal structure of BAK core domain BH3-groove-dimer in complex with POPC and C8E4 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-ETHANEDIOL, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, ... | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-11-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | BAK core dimers bind lipids and can be bridged by them.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VBZ
 
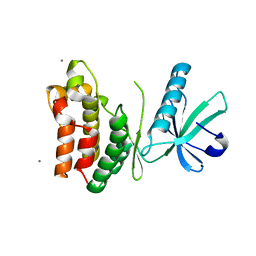 | | Crystal structure of the rat MLKL pseudokinase domain | | Descriptor: | MANGANESE (II) ION, Mixed lineage kinase domain-like pseudokinase | | Authors: | Davies, K.A, Czabotar, P.E. | | Deposit date: | 2019-12-19 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | Distinct pseudokinase domain conformations underlie divergent activation mechanisms among vertebrate MLKL orthologues.
Nat Commun, 11, 2020
|
|
6UXN
 
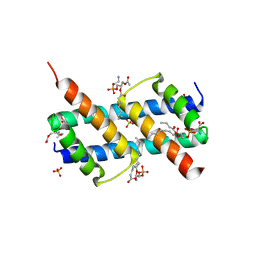 | | Crystal structure of BAK core domain BH3-groove-dimer in complex with phosphatidylserine | | Descriptor: | Bcl-2 homologous antagonist/killer, GLYCEROL, O-[(R)-{[(2R)-2,3-bis(octanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, ... | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-11-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | BAK core dimers bind lipids and can be bridged by them.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6UXP
 
 | | Crystal structure of BAK core domain BH3-groove-dimer in complex with phosphatidylglycerol | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-[PHOSPHO-RAC-(1-GLYCEROL), Bcl-2 homologous antagonist/killer, GLYCEROL | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-11-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | BAK core dimers bind lipids and can be bridged by them.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6UXM
 
 | |
6UXR
 
 | | Crystal structure of BAK core domain BH3-groove-dimer in complex with LysoPC | | Descriptor: | Bcl-2 homologous antagonist/killer, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-11-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | BAK core dimers bind lipids and can be bridged by them.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VC0
 
 | | Crystal structure of the horse MLKL pseudokinase domain | | Descriptor: | GLYCEROL, Mixed lineage kinase domain like pseudokinase | | Authors: | Davies, K.A, Czabotar, P.E. | | Deposit date: | 2019-12-19 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.746 Å) | | Cite: | Distinct pseudokinase domain conformations underlie divergent activation mechanisms among vertebrate MLKL orthologues.
Nat Commun, 11, 2020
|
|
6UVC
 
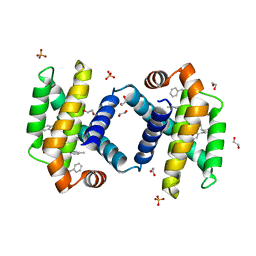 | | Crystal structure of BCL-XL bound to compound 8: (R)-3-(Benzylthio)-2-(3-(2-((4'-chloro-[1,1'-biphenyl]-2-yl)methyl)-1,2,3,4-tetrahydroisoquinoline-6-carbonyl)-3-(4-methylbenzyl)ureido)propanoic acid | | Descriptor: | (R)-3-(Benzylthio)-2-(3-(2-((4'-chloro-[1,1'-biphenyl]-2-yl)methyl)-1,2,3,4-tetrahydroisoquinoline-6-carbonyl)-3-(4-methylbenzyl)ureido)propanoic acid, 1,2-ETHANEDIOL, Bcl-2-like protein 1, ... | | Authors: | Roy, M.J, Birkinshaw, R, Lessene, G, Czabotar, P.E. | | Deposit date: | 2019-11-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Development of Potent Benzoylurea Inhibitors of BCL-X L and BCL-2.
J.Med.Chem., 64, 2021
|
|
6UVE
 
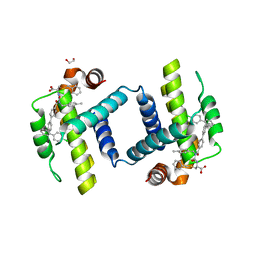 | | Crystal structure of BCL-XL bound to compound 7: (R)-3-(Benzylthio)-2-(3-(4-chloro-[1,1':2',1'':3'',1'''-quaterphenyl]-4'''-carbonyl)-3-(4-methylbenzyl)ureido)propanoic acid | | Descriptor: | (R)-3-(Benzylthio)-2-(3-(4-chloro-[1,1':2',1'':3'',1'''-quaterphenyl]-4'''-carbonyl)-3-(4-methylbenzyl)ureido)propanoic acid, 1,2-ETHANEDIOL, Bcl-2-like protein 1 | | Authors: | Roy, M.J, Lessene, G, Czabotar, P.E. | | Deposit date: | 2019-11-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structure-Guided Development of Potent Benzoylurea Inhibitors of BCL-X L and BCL-2.
J.Med.Chem., 64, 2021
|
|
6UVD
 
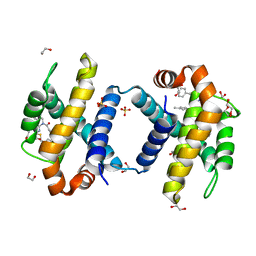 | | Crystal structure of BCL-XL bound to compound 2: (2R)-3-(Benzylsulfanyl)-2-({[(4-methylphenyl)methyl] [(4 phenylphenyl)carbonyl] carbamoyl}amino) propanoic acid | | Descriptor: | (2R)-3-(Benzylsulfanyl)-2-({[(4-methylphenyl)methyl] [(4 phenylphenyl)carbonyl] carbamoyl}amino) propanoic acid, 1,2-ETHANEDIOL, Bcl-2-like protein 1, ... | | Authors: | Roy, M.J, Birkinshaw, R, Lessene, G, Czabotar, P.E. | | Deposit date: | 2019-11-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Guided Development of Potent Benzoylurea Inhibitors of BCL-X L and BCL-2.
J.Med.Chem., 64, 2021
|
|
6UVH
 
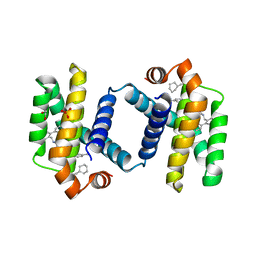 | | Crystal structure of BCL-XL bound to compound 15: (R)-2-(3-(2-((4'-Chloro-[1,1'-biphenyl]-2-yl)methyl)-1,2,3,4-tetrahydroisoquinoline-6-carbonyl)-3-(4-methylbenzyl)ureido)-3-((cyclohexylmethyl)sulfonyl)propanoic acid | | Descriptor: | (R)-2-(3-(2-((4'-Chloro-[1,1'-biphenyl]-2-yl)methyl)-1,2,3,4-tetrahydroisoquinoline-6-carbonyl)-3-(4-methylbenzyl)ureido)-3-((cyclohexylmethyl)sulfonyl)propanoic acid, 1,2-ETHANEDIOL, Bcl-2-like protein 1, ... | | Authors: | Roy, M.J, Lessene, G, Czabotar, P.E. | | Deposit date: | 2019-11-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure-Guided Development of Potent Benzoylurea Inhibitors of BCL-X L and BCL-2.
J.Med.Chem., 64, 2021
|
|
6UVF
 
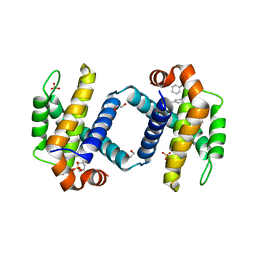 | | Crystal structure of BCL-XL bound to compound 12: (R)-2-(3-([1,1'-Biphenyl]-4-carbonyl)-3-(4-methylbenzyl)ureido)-3-((cyclohexylmethyl)sulfonyl)propanoic acid | | Descriptor: | (R)-2-(3-([1,1'-Biphenyl]-4-carbonyl)-3-(4-methylbenzyl)ureido)-3-((cyclohexylmethyl)sulfonyl)propanoic acid, 1,2-ETHANEDIOL, Bcl-2-like protein 1, ... | | Authors: | Roy, M.J, Lessene, G, Czabotar, P.E. | | Deposit date: | 2019-11-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure-Guided Development of Potent Benzoylurea Inhibitors of BCL-X L and BCL-2.
J.Med.Chem., 64, 2021
|
|
