3ICD
 
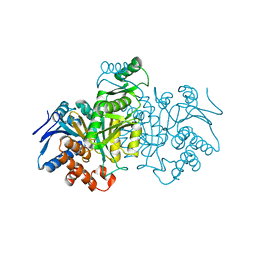 | | STRUCTURE OF A BACTERIAL ENZYME REGULATED BY PHOSPHORYLATION, ISOCITRATE DEHYDROGENASE | | Descriptor: | ISOCITRATE DEHYDROGENASE | | Authors: | Hurley, J.H, Thorsness, P.E, Ramalingam, V, Helmers, N.H, Koshlandjunior, D.E, Stroud, R.M. | | Deposit date: | 1989-12-28 | | Release date: | 1991-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a bacterial enzyme regulated by phosphorylation, isocitrate dehydrogenase.
Proc.Natl.Acad.Sci.USA, 86, 1989
|
|
5L82
 
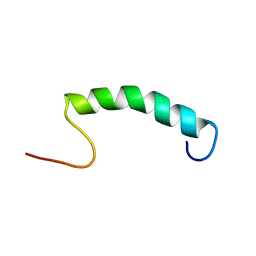 | |
5LFI
 
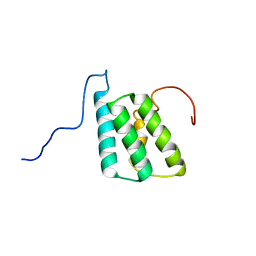 | | lactococcin A immunity protein | | Descriptor: | Lactococcin-A immunity protein | | Authors: | Persson, C, Fuochi, V, Pedersen, A, Karlsson, B.G, Nissen-Meyer, J, Kristiansen, P.E, Oppegard, C. | | Deposit date: | 2016-07-01 | | Release date: | 2016-11-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Structure and Mutational Analysis of the Lactococcin A Immunity Protein.
Biochemistry, 55, 2016
|
|
5MMS
 
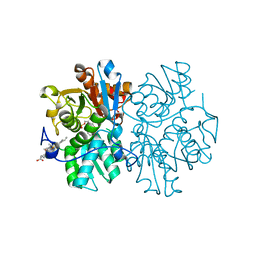 | | Human cystathionine beta-synthase (CBS) p.P49L delta409-551 variant | | Descriptor: | Cystathionine beta-synthase, PROTOPORPHYRIN IX CONTAINING FE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Vicente, J.B, Colaco, H.G, Malagrino, F, Santo, P.E, Gutierres, A, Bandeiras, T.M, Leandro, P, Brito, J.A, Giuffre, A. | | Deposit date: | 2016-12-12 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Clinically Relevant Variant of the Human Hydrogen Sulfide-Synthesizing Enzyme Cystathionine beta-Synthase: Increased CO Reactivity as a Novel Molecular Mechanism of Pathogenicity?
Oxid Med Cell Longev, 2017, 2017
|
|
5MYG
 
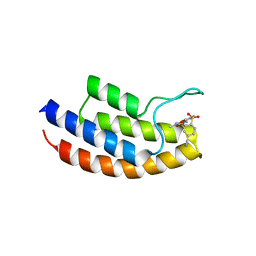 | | Crystal structure of the bromodomain of human BRPF1 in complex with NI-57 chemical probe | | Descriptor: | 4-cyano-~{N}-(1,3-dimethyl-2-oxidanylidene-quinolin-6-yl)-2-methoxy-benzenesulfonamide, Peregrin | | Authors: | Tallant, C, Igoe, N, Bayle, E.D, Krojer, T, Nunez-Alonso, G, Kopec, J, Fitzpatrick, F, Savitsky, P, Fedorov, O, Brennan, P.E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Muller, S, Fish, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-26 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of a Chemical Probe for the Bromodomain and Plant Homeodomain Finger-Containing (BRPF) Family of Proteins.
J. Med. Chem., 60, 2017
|
|
5NQ4
 
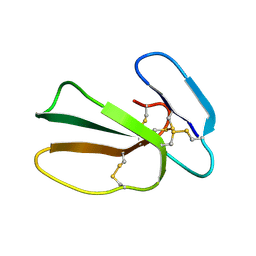 | | Cytotoxin-1 in DPC-micelle | | Descriptor: | Cytotoxin 1 | | Authors: | Dubovskii, P.V, Dubinnyi, M.A, Volynsky, P.E, Pustovalova, Y.E, Konshina, A.G, Utkin, Y.N, Efremov, R.G, Arseniev, A.S. | | Deposit date: | 2017-04-19 | | Release date: | 2017-12-13 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Impact of membrane partitioning on the spatial structure of an S-type cobra cytotoxin.
J. Biomol. Struct. Dyn., 36, 2018
|
|
8G1T
 
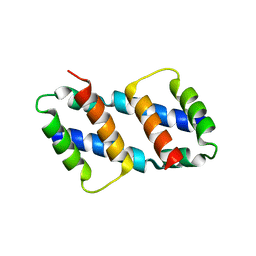 | | Crystal structure of Bax core domain BH3-groove dimer - tetrameric fraction P21 | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BAX | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E, Miller, M.S. | | Deposit date: | 2023-02-02 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 2023
|
|
8GHR
 
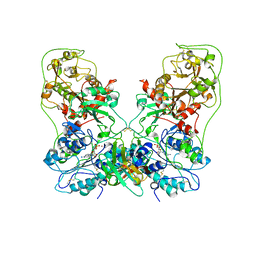 | | Structure of human ENPP1 in complex with variable heavy domain VH27.2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Carozza, J.A, Wang, H, Solomon, P.E, Wells, J.A, Li, L. | | Deposit date: | 2023-03-10 | | Release date: | 2023-08-02 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Discovery of VH domains that allosterically inhibit ENPP1.
Nat.Chem.Biol., 20, 2024
|
|
1CX1
 
 | | SECOND N-TERMINAL CELLULOSE-BINDING DOMAIN FROM CELLULOMONAS FIMI BETA-1,4-GLUCANASE C, NMR, 22 STRUCTURES | | Descriptor: | ENDOGLUCANASE C | | Authors: | Brun, E, Johnson, P.E, Creagh, L.A, Haynes, C.A, Tomme, P, Webster, P, Kilburn, D.G, McIntosh, L.P. | | Deposit date: | 1999-08-27 | | Release date: | 2000-04-02 | | Last modified: | 2017-02-01 | | Method: | SOLUTION NMR | | Cite: | Structure and binding specificity of the second N-terminal cellulose-binding domain from Cellulomonas fimi endoglucanase C.
Biochemistry, 39, 2000
|
|
8BNA
 
 | | BINDING OF HOECHST 33258 TO THE MINOR GROOVE OF B-DNA | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Pjura, P.E, Grzeskowiak, K, Dickerson, R.E. | | Deposit date: | 1986-08-29 | | Release date: | 1987-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding of Hoechst 33258 to the minor groove of B-DNA.
J.Mol.Biol., 197, 1987
|
|
1TF3
 
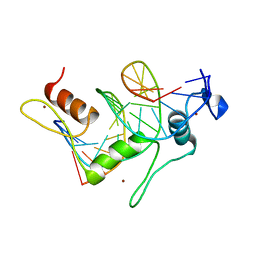 | | TFIIIA FINGER 1-3 BOUND TO DNA, NMR, 22 STRUCTURES | | Descriptor: | 5S RNA GENE, TRANSCRIPTION FACTOR IIIA, ZINC ION | | Authors: | Foster, M.P, Wuttke, D.S, Radhakrishnan, I, Case, D.A, Gottesfeld, J.M, Wright, P.E. | | Deposit date: | 1997-07-01 | | Release date: | 1997-09-17 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Domain packing and dynamics in the DNA complex of the N-terminal zinc fingers of TFIIIA.
Nat.Struct.Biol., 4, 1997
|
|
1ULP
 
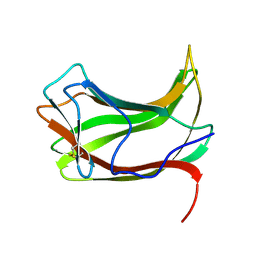 | |
1ULO
 
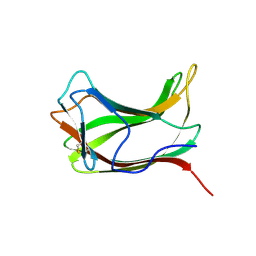 | |
1XJ9
 
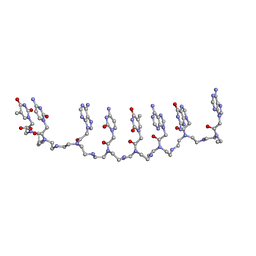 | | Crystal structure of a partly self-complementary peptide nucleic acid (PNA) oligomer showing a duplex-triplex network | | Descriptor: | peptide nucleic acid, (H-P(*GPN*TPN*APN*GPN*APN*TPN*CPN*APN*CPN*TPN)-LYS-NH2) | | Authors: | Petersson, B, Nielsen, B.B, Rasmussen, H, Larsen, I.K, Gajhede, M, Nielsen, P.E, Kastrup, J.S. | | Deposit date: | 2004-09-23 | | Release date: | 2005-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a Partly Self-Complementary Peptide Nucleic Acid (PNA) Oligomer Showing a Duplex-Triplex Network
J.Am.Chem.Soc., 127, 2005
|
|
3UBI
 
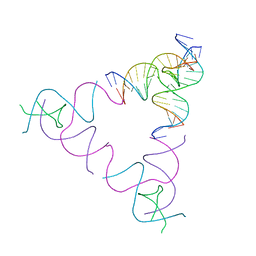 | | The Absence of Tertiary Interactions in a Self-Assembled DNA Crystal Structure | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*CP*GP*TP*AP*CP*TP*CP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*TP*GP*AP*GP*GP*CP*TP*GP*C)-3'), DNA (5'-D(P*CP*CP*GP*AP*GP*TP*AP*CP*GP*AP*CP*GP*AP*CP*AP*AP*G)-3'), ... | | Authors: | Nguyen, N, Birktoft, J.J, Sha, R, Wang, T, Zheng, J, Constantinou, P.E, Ginell, S.L, Chen, Y, Mao, C, Seeman, N.C. | | Deposit date: | 2011-10-24 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (6.8046 Å) | | Cite: | The absence of tertiary interactions in a self-assembled DNA crystal structure.
J.Mol.Recognit., 25, 2012
|
|
1RRU
 
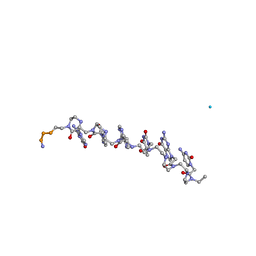 | | The influence of a chiral amino acid on the helical handedness of PNA in solution and in crystals | | Descriptor: | Peptide Nucleic Acid, (H-P(*CPN*GPN*TPN*APN*CPN*GPN)-LYS-NH2) | | Authors: | Rasmussen, H, Liljefors, T, Petersson, B, Nielsen, P.E, Kastrup, J.S. | | Deposit date: | 2003-12-09 | | Release date: | 2004-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Influence of a Chiral Amino Acid on the Helical Handedness of PNA in Solution and in Crystals
J.Biomol.Struct.Dyn., 21, 2004
|
|
3D7V
 
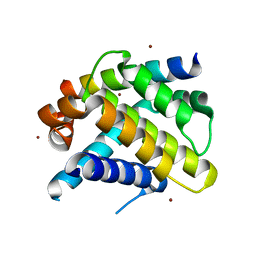 | | Crystal structure of Mcl-1 in complex with an Mcl-1 selective BH3 ligand | | Descriptor: | Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Lee, E.F, Czabotar, P.E, Colman, P.M, Fairlie, W.D. | | Deposit date: | 2008-05-22 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A novel BH3 ligand that selectively targets Mcl-1 reveals that apoptosis can proceed without Mcl-1 degradation.
J.Cell Biol., 180, 2008
|
|
3DKS
 
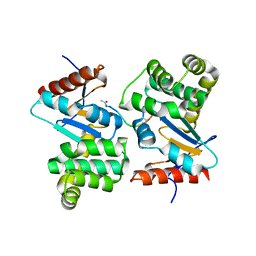 | | DsbA substrate complex | | Descriptor: | Thiol:disulfide interchange protein dsbA, siga peptide | | Authors: | Paxman, J.J, Borg, N.A, Horne, J, Rossjohn, J, Thompson, P.E, Piek, S, Kahler, C.M, Sakellaris, H, Scanlon, M.J. | | Deposit date: | 2008-06-25 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of the bacterial oxidoreductase enzyme DsbA in complex with a peptide reveals a basis for substrate specificity in the catalytic cycle of DsbA enzymes
J.Biol.Chem., 284, 2009
|
|
3FDL
 
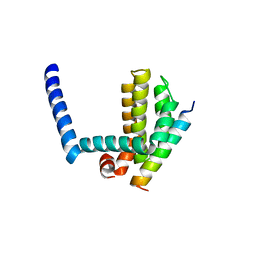 | | Bim BH3 peptide in complex with Bcl-xL | | Descriptor: | Apoptosis regulator Bcl-X, Bcl-2-like protein 11 | | Authors: | Fairlie, W.D, Lee, E.F, Smith, B.J, Czabotar, P.E, Colman, P.M. | | Deposit date: | 2008-11-26 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | High-Resolution Structural Characterization of a Helical alpha/beta-Peptide Foldamer Bound to the Anti-Apoptotic Protein Bcl-x(L)
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3FDM
 
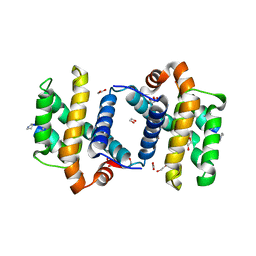 | | alpha/beta foldamer in complex with Bcl-xL | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator Bcl-X, alpha/beta-peptide foldamer | | Authors: | Fairlie, W.D, Lee, E.F, Smith, B.J, Czabotar, P.E, Colman, P.M, Sadowsky, J.D, Peterson-Kaufman, K.J, Gellman, S.H. | | Deposit date: | 2008-11-26 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | High-Resolution Structural Characterization of a Helical alpha/beta-Peptide Foldamer Bound to the Anti-Apoptotic Protein Bcl-x(L)
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
7AV9
 
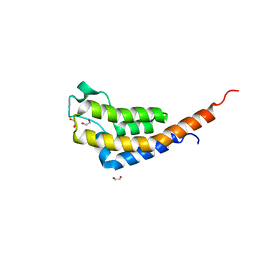 | | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group C2 | | Descriptor: | 1,2-ETHANEDIOL, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group C2
To Be Published
|
|
7B3J
 
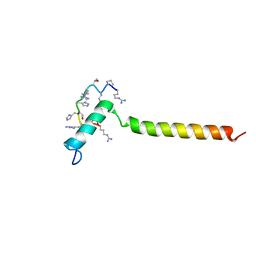 | | Dynamic complex between all-D-enantiomeric peptide D3 with wild-type amyloid precursor protein 672-726 fragment (amyloid beta 1-55) | | Descriptor: | D3 all D-enantimeric peptide, Isoform L-APP677 of Amyloid-beta precursor protein | | Authors: | Bocharov, E.V, Volynsky, P.E, Okhrimenko, I.S, Urban, A.S. | | Deposit date: | 2020-12-01 | | Release date: | 2021-01-13 | | Last modified: | 2021-12-08 | | Method: | SOLUTION NMR | | Cite: | All - d - Enantiomeric Peptide D3 Designed for Alzheimer's Disease Treatment Dynamically Interacts with Membrane-Bound Amyloid-beta Precursors.
J.Med.Chem., 64, 2021
|
|
7AV8
 
 | | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group P21212 | | Descriptor: | PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group P21212
To Be Published
|
|
7B3K
 
 | | Dynamic complex between all-D-enantiomeric peptide D3 with L723P mutant of amyloid precursor protein (APP) 672-726 fragment (amyloid beta 1-55) | | Descriptor: | D3 all D-enantimeric peptide, Isoform L-APP677 of Amyloid-beta precursor protein | | Authors: | Bocharov, E.V, Volynsky, P.E, Okhrimenko, I.S, Urban, A.S. | | Deposit date: | 2020-12-01 | | Release date: | 2021-01-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | All - d - Enantiomeric Peptide D3 Designed for Alzheimer's Disease Treatment Dynamically Interacts with Membrane-Bound Amyloid-beta Precursors.
J.Med.Chem., 64, 2021
|
|
7BBO
 
 | | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group P212121 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group P21212
To Be Published
|
|
