1M5A
 
 | |
8CXC
 
 | | Novel Anti-Mesothelin Antibodies Enable Crystallography of the Intact Mesothelin Ectodo- main and Engineering of Potent, T cell-engaging Bispecific Therapeutics | | Descriptor: | 3F2 Antibody heavy chain, 3F2 Antibody light chain, Mesothelin, ... | | Authors: | Bandaranayake, A.D, Rupert, P.B, Lin, I, Pilat, K, Ruff, R.O, Friend, D.J, Chan, M.K, Clarke, M, Carter, J, Meshinchi, S, Mehlin, C, Olson, J.M, Strong, R.K, Correnti, C.E. | | Deposit date: | 2022-05-20 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.31 Å) | | Cite: | Novel mesothelin antibodies enable crystallography of the intact mesothelin ectodomain and engineering of potent, T cell-engaging bispecific therapeutics
Front. Drug Discov., 3, 2023
|
|
8CYH
 
 | | Novel Anti-Mesothelin Antibodies Enable Crystallography of the Intact Mesothelin Ectodo- main and Engineering of Potent, T cell-engaging Bispecific Therapeutics | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, A12 antibody heavy chain, A12 antibody light chain, ... | | Authors: | Bandaranayake, A.D, Rupert, P.B, Lin, I, Pilat, K, Ruff, R.O, Friend, D.J, Chan, M.K, Clarke, M, Carter, J, Meshinchi, S, Mehlin, C, Olson, J.M, Strong, R.K, Correnti, C.E. | | Deposit date: | 2022-05-23 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Novel mesothelin antibodies enable crystallography of the intact mesothelin ectodomain and engineering of potent, T cell-engaging bispecific therapeutics
Front. Drug Discov., 3, 2023
|
|
8CZ8
 
 | | Novel Anti-Mesothelin Antibodies Enable Crystallography of the Intact Mesothelin Ectodo- main and Engineering of Potent, T cell-engaging Bispecific Therapeutics | | Descriptor: | Mesothelin, cleaved form, SULFATE ION, ... | | Authors: | Bandaranayake, A.D, Rupert, P.B, Lin, I, Pilat, K, Ruff, R.O, Friend, D.J, Chan, M.K, Clarke, M, Carter, J, Meshinchi, S, Mehlin, C, Olson, J.M, Strong, R.K, Correnti, C.E. | | Deposit date: | 2022-05-24 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel mesothelin antibodies enable crystallography of the intact mesothelin ectodomain and engineering of potent, T cell-engaging bispecific therapeutics
Front. Drug Discov., 3, 2023
|
|
1HQ3
 
 | | CRYSTAL STRUCTURE OF THE HISTONE-CORE-OCTAMER IN KCL/PHOSPHATE | | Descriptor: | CHLORIDE ION, HISTONE H2A-IV, HISTONE H2B, ... | | Authors: | Chantalat, L, Nicholson, J.M, Lambert, S.J, Reid, A.J, Donovan, M.J, Reynolds, C.D, Wood, C.M, Baldwin, J.P. | | Deposit date: | 2000-12-14 | | Release date: | 2001-01-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the histone-core octamer in KCl/phosphate crystals at 2.15 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2ARO
 
 | | Crystal Structure Of The Native Histone Octamer To 2.1 Angstrom Resolution, Crystalised In The Presence Of S-Nitrosoglutathione | | Descriptor: | CHLORIDE ION, HISTONE H3, HISTONE H4-VI, ... | | Authors: | Wood, C.M, Sodngam, S, Nicholson, J.M, Lambert, S.J, Reynolds, C.D, Baldwin, J.P. | | Deposit date: | 2005-08-20 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The oxidised histone octamer does not form a H3 disulphide bond.
Biochim.Biophys.Acta, 1764, 2006
|
|
1TZY
 
 | | Crystal Structure of the Core-Histone Octamer to 1.90 Angstrom Resolution | | Descriptor: | CHLORIDE ION, HISTONE H3, HISTONE H4-VI, ... | | Authors: | Wood, C.M, Nicholson, J.M, Chantalat, L, Reynolds, C.D, Lambert, S.J, Baldwin, J.P. | | Deposit date: | 2004-07-12 | | Release date: | 2004-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution structure of the native histone octamer.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
5JG9
 
 | |
6AVC
 
 | |
6ATU
 
 | |
6ATS
 
 | |
6ATL
 
 | | Exploring Cystine Dense Peptide Space to Open a Unique Molecular Toolbox | | Descriptor: | CITRIC ACID, Potassium channel toxin alpha-KTx 4.2, SULFATE ION | | Authors: | Gewe, M.M, Rupert, P, Strong, R.K. | | Deposit date: | 2017-08-29 | | Release date: | 2018-02-28 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Screening, large-scale production and structure-based classification of cystine-dense peptides.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6AUP
 
 | | Exploring Cystine Dense Peptide Space to Open a Unique Molecular Toolbox | | Descriptor: | GLYCEROL, Potassium channel toxin gamma-KTx 2.2, SULFATE ION | | Authors: | Gewe, M.M, Rupert, P, Strong, R.K. | | Deposit date: | 2017-09-01 | | Release date: | 2018-02-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Screening, large-scale production and structure-based classification of cystine-dense peptides.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6ATN
 
 | |
6AVD
 
 | |
4BCL
 
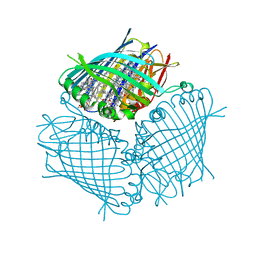 | | FMO protein from Prosthecochloris aestuarii 2K at Room Temperature | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOCHLOROPHYLL A PROTEIN | | Authors: | Tronrud, D.E, Matthews, B.W. | | Deposit date: | 1998-04-17 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Refinement of the Structure of a Water-Soluble Antenna Complex from Green Photosynthetic Bacteria by Incorporation of the Chemically Determined Amino Acid Sequence
Photosynthetic Reaction Center, 1, 1993
|
|
3EOJ
 
 | | Fmo protein from Prosthecochloris Aestuarii 2K AT 1.3A Resolution | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, BACTERIOCHLOROPHYLL A, ... | | Authors: | Tronrud, D.E, Wen, J, Gay, L, Blankenship, R.E. | | Deposit date: | 2008-09-27 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structural basis for the difference in absorbance spectra for the FMO antenna protein from various green sulfur bacteria.
Photosynth.Res., 100, 2009
|
|
7SND
 
 | | Pacifastin related protease inhibitors | | Descriptor: | GLYCEROL, PHOSPHATE ION, Pacifastin-related peptide | | Authors: | Gewe, M.M, Strong, R.K. | | Deposit date: | 2021-10-27 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Ex silico engineering of cystine-dense peptides yielding a potent bispecific T cell engager.
Sci Transl Med, 14, 2022
|
|
7SNC
 
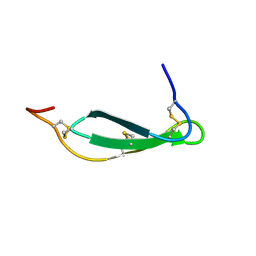 | | Pacifastin related protease inhibitors | | Descriptor: | Protease inhibitor | | Authors: | Gewe, M.M, Strong, R.K. | | Deposit date: | 2021-10-27 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ex silico engineering of cystine-dense peptides yielding a potent bispecific T cell engager.
Sci Transl Med, 14, 2022
|
|
5JHI
 
 | | Solution structure of the de novo mini protein gEHE_06 | | Descriptor: | W35 | | Authors: | Buchko, G.W, Bahl, C.D, Gilmore, J.M, Pulavarti, S.V, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-21 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
5JI4
 
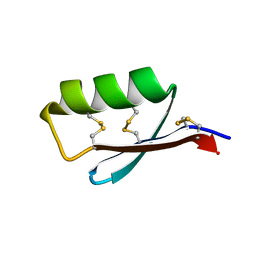 | | Solution structure of the de novo mini protein gEEHE_02 | | Descriptor: | W37 | | Authors: | Buchko, G.W, Bahl, C.D, Pulavarti, S.V, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-21 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
5KWX
 
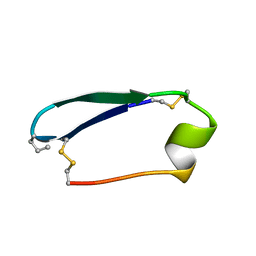 | |
5KWO
 
 | |
5KWZ
 
 | |
5KX2
 
 | |
