2RT9
 
 | | Solution structure of a regulatory domain of meiosis inhibitor | | 分子名称: | F-box only protein 43, ZINC ION | | 著者 | Shoji, S, Muto, Y, Ikeda, M, He, F, Tsuda, K, Ohsawa, N, Akasaka, R, Terada, T, Wakiyama, M, Shirouzu, M, Yokoyama, S. | | 登録日 | 2013-07-05 | | 公開日 | 2014-07-16 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The zinc-binding region (ZBR) fragment of Emi2 can inhibit APC/C by targeting its association with the coactivator Cdc20 and UBE2C-mediated ubiquitylation
FEBS Open Bio, 4, 2014
|
|
3A8Y
 
 | | Crystal structure of the complex between the BAG5 BD5 and Hsp70 NBD | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BAG family molecular chaperone regulator 5, Heat shock 70 kDa protein 1 | | 著者 | Arakawa, A, Handa, N, Ohsawa, N, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2009-10-13 | | 公開日 | 2010-03-31 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The C-terminal BAG domain of BAG5 induces conformational changes of the Hsp70 nucleotide-binding domain for ADP-ATP exchange
Structure, 18, 2010
|
|
3VUZ
 
 | | Crystal structure of histone methyltransferase SET7/9 in complex with AAM-1 | | 分子名称: | 5'-{[(3S)-3-amino-3-carboxypropyl](hexyl)amino}-5'-deoxyadenosine, Histone-lysine N-methyltransferase SETD7 | | 著者 | Niwa, H, Handa, N, Tomabechi, Y, Honda, K, Toyama, M, Ohsawa, N, Shirouzu, M, Kagechika, H, Hirano, T, Umehara, T, Yokoyama, S. | | 登録日 | 2012-07-10 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of histone methyltransferase SET7/9 in complexes with adenosylmethionine derivatives
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VV0
 
 | | Crystal structure of histone methyltransferase SET7/9 in complex with DAAM-3 | | 分子名称: | 5'-{[(3S)-3-amino-3-carboxypropyl][2-(hexylamino)ethyl]amino}-5'-deoxyadenosine, Histone-lysine N-methyltransferase SETD7 | | 著者 | Niwa, H, Handa, N, Tomabechi, Y, Honda, K, Toyama, M, Ohsawa, N, Shirouzu, M, Kagechika, H, Hirano, T, Umehara, T, Yokoyama, S. | | 登録日 | 2012-07-10 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Structures of histone methyltransferase SET7/9 in complexes with adenosylmethionine derivatives
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2YXM
 
 | | Crystal structure of I-set domain of human Myosin Binding ProteinC | | 分子名称: | Myosin-binding protein C, slow-type | | 著者 | Kishishita, S, Ohsawa, N, Murayama, K, Chen, L, Liu, Z, Terada, T, Shirouzu, M, Wang, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-04-26 | | 公開日 | 2007-10-30 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Crystal structure of I-set domain of human Myosin Binding ProteinC
To be Published
|
|
2YZ8
 
 | | Crystal structure of the 32th Ig-like domain of human obscurin (KIAA1556) | | 分子名称: | Obscurin | | 著者 | Saijo, S, Ohsawa, N, Nishino, A, Kishishita, S, Chen, L, Fu, Z.Q, Chrzas, J, Wang, B.C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-05-04 | | 公開日 | 2008-05-06 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the 32th Ig-like domain of human obscurin (KIAA1556)
To be Published
|
|
3VR2
 
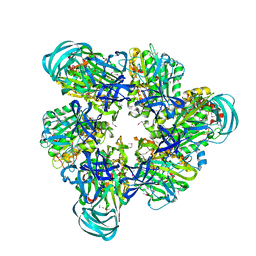 | | Crystal structure of nucleotide-free A3B3 complex from Enterococcus hirae V-ATPase [eA3B3] | | 分子名称: | V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B | | 著者 | Arai, S, Saijo, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | 登録日 | 2012-04-03 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
3VR4
 
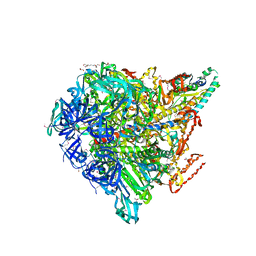 | | Crystal structure of Enterococcus hirae V1-ATPase [eV1] | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | 著者 | Saijo, S, Arai, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | 登録日 | 2012-04-03 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.172 Å) | | 主引用文献 | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
3VR6
 
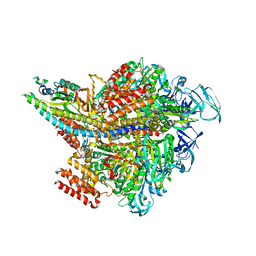 | | Crystal structure of AMP-PNP bound Enterococcus hirae V1-ATPase [bV1] | | 分子名称: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, V-type sodium ATPase catalytic subunit A, ... | | 著者 | Arai, S, Saijo, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | 登録日 | 2012-04-03 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
3VR3
 
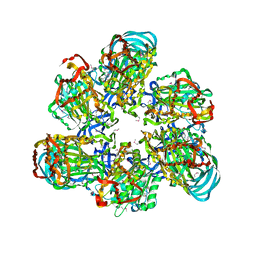 | | Crystal structure of AMP-PNP bound A3B3 complex from Enterococcus hirae V-ATPase [bA3B3] | | 分子名称: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, V-type sodium ATPase catalytic subunit A, ... | | 著者 | Arai, S, Saijo, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | 登録日 | 2012-04-03 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
3VR5
 
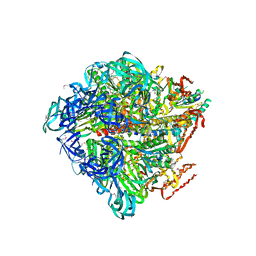 | | Crystal structure of nucleotide-free Enterococcus hirae V1-ATPase [eV1(L)] | | 分子名称: | V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B, V-type sodium ATPase subunit D, ... | | 著者 | Saijo, S, Arai, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | 登録日 | 2012-04-03 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (3.9 Å) | | 主引用文献 | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
4YN3
 
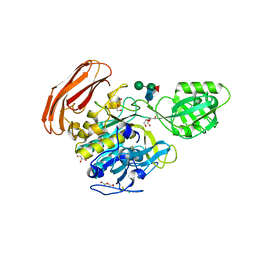 | | Crystal structure of Cucumisin complex with pro-peptide | | 分子名称: | CHLORIDE ION, Cucumisin, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Murayama, K, Kato-Murayama, M, Yokoyama, S, Arima, K, Shirouzu, M. | | 登録日 | 2015-03-09 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural basis of cucumisin protease activity regulation by its propeptide
J. Biochem., 161, 2017
|
|
2RQR
 
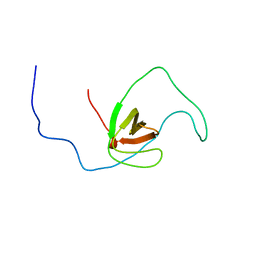 | | The solution structure of human DOCK2 SH3 domain - ELMO1 peptide chimera complex | | 分子名称: | Engulfment and cell motility protein 1,Dedicator of cytokinesis protein 2 | | 著者 | Yokoyama, S, Tochio, N, Koshiba, S, Kigawa, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2009-10-21 | | 公開日 | 2010-10-27 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for mutual relief of the Rac guanine nucleotide exchange factor DOCK2 and its partner ELMO1 from their autoinhibited forms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5AX0
 
 | | Crystal Structure of the Cell-Free Synthesized Membrane Protein, Acetabularia Rhodopsin I, at 1.52 angstrom | | 分子名称: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DECANE, DODECANE, ... | | 著者 | Furuse, M, Hosaka, T, Kimura-Someya, T, Yokoyama, S, Shirouzu, M. | | 登録日 | 2015-07-10 | | 公開日 | 2015-08-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.521 Å) | | 主引用文献 | Structural basis for the slow photocycle and late proton release in Acetabularia rhodopsin I from the marine plant Acetabularia acetabulum
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5AWZ
 
 | | Crystal Structure of the Cell-Free Synthesized Membrane Protein, Acetabularia Rhodopsin I, at 1.57 angstrom | | 分子名称: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DECANE, DODECANE, ... | | 著者 | Furuse, M, Hosaka, T, Kimura-Someya, T, Yokoyama, S, Shirouzu, M. | | 登録日 | 2015-07-10 | | 公開日 | 2015-08-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Structural basis for the slow photocycle and late proton release in Acetabularia rhodopsin I from the marine plant Acetabularia acetabulum
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5AX1
 
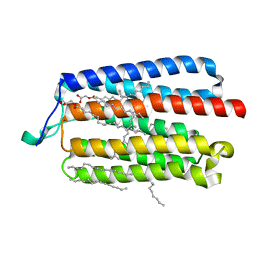 | | Crystal Structure of the Cell-Free Synthesized Membrane Protein, Acetabularia Rhodopsin I, at 1.80 angstrom | | 分子名称: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DECANE, DODECANE, ... | | 著者 | Furuse, M, Hosaka, T, Kimura-Someya, T, Yokoyama, S, Shirouzu, M. | | 登録日 | 2015-07-10 | | 公開日 | 2015-08-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.803 Å) | | 主引用文献 | Structural basis for the slow photocycle and late proton release in Acetabularia rhodopsin I from the marine plant Acetabularia acetabulum
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5Y7Z
 
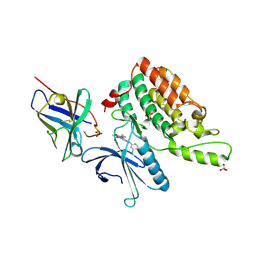 | | Complex structure of cyclin G-associated kinase with gefitinib | | 分子名称: | Cyclin-G-associated kinase, Gefitinib, NANOBODY, ... | | 著者 | Ohbayashi, N, Murayama, K, Kato-Murayama, M, Shirouzu, M. | | 登録日 | 2017-08-18 | | 公開日 | 2018-08-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.804 Å) | | 主引用文献 | Structural Basis for the Inhibition of Cyclin G-Associated Kinase by Gefitinib.
ChemistryOpen, 7, 2018
|
|
5Y80
 
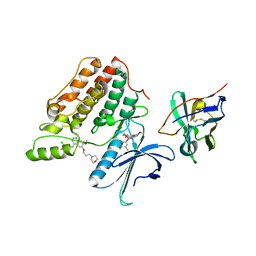 | | Complex structure of cyclin G-associated kinase with gefitinib | | 分子名称: | Cyclin-G-associated kinase, Gefitinib, NANOBODY | | 著者 | Ohbayashi, N, Murayama, K, Kato-Murayama, M, Shirouzu, M. | | 登録日 | 2017-08-18 | | 公開日 | 2018-08-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Basis for the Inhibition of Cyclin G-Associated Kinase by Gefitinib.
ChemistryOpen, 7, 2018
|
|
5XAX
 
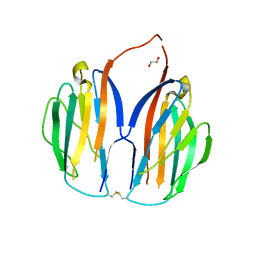 | |
5XAW
 
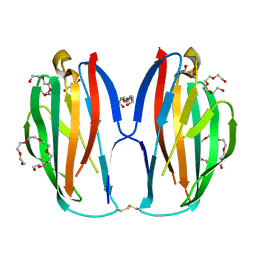 | | Parallel homodimer structures of voltage-gated sodium channel beta4 for cell-cell adhesion | | 分子名称: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, GLYCEROL, Sodium channel subunit beta-4, ... | | 著者 | Shimizu, H, Yokoyama, S. | | 登録日 | 2017-03-15 | | 公開日 | 2017-07-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.101 Å) | | 主引用文献 | Parallel homodimer structures of the extracellular domains of the voltage-gated sodium channel beta 4 subunit explain its role in cell-cell adhesion
J. Biol. Chem., 292, 2017
|
|
6AJ4
 
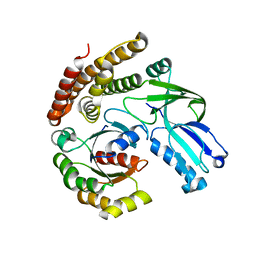 | |
6AJL
 
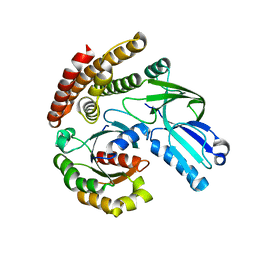 | | DOCK7 mutant I1836Y complexed with Cdc42 | | 分子名称: | Cell division control protein 42 homolog, Dedicator of cytokinesis protein 7 | | 著者 | Kukimoto-Niino, M, Shirouzu, M. | | 登録日 | 2018-08-28 | | 公開日 | 2019-03-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.23 Å) | | 主引用文献 | Structural Basis for the Dual Substrate Specificity of DOCK7 Guanine Nucleotide Exchange Factor.
Structure, 27, 2019
|
|
1UGO
 
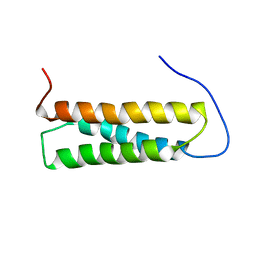 | | Solution structure of the first Murine BAG domain of Bcl2-associated athanogene 5 | | 分子名称: | Bcl2-associated athanogene 5 | | 著者 | Endoh, H, Hayashi, F, Seimiya, K, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-06-17 | | 公開日 | 2004-08-03 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The C-terminal BAG domain of BAG5 induces conformational changes of the Hsp70 nucleotide-binding domain for ADP-ATP exchange
Structure, 18, 2010
|
|
1UK5
 
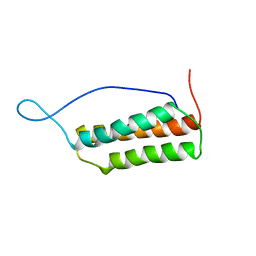 | | Solution structure of the Murine BAG domain of Bcl2-associated athanogene 3 | | 分子名称: | BAG-family molecular chaperone regulator-3 | | 著者 | Hatta, R, Yoshida, M, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-08-19 | | 公開日 | 2004-02-19 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The C-terminal BAG domain of BAG5 induces conformational changes of the Hsp70 nucleotide-binding domain for ADP-ATP exchange
Structure, 18, 2010
|
|
5B2G
 
 | | Crystal structure of human claudin-4 in complex with C-terminal fragment of Clostridium perfringens enterotoxin | | 分子名称: | Endolysin,Claudin-4, Heat-labile enterotoxin B chain | | 著者 | Shinoda, T, Kimura-Someya, T, Shirouzu, M, Yokoyama, S. | | 登録日 | 2016-01-15 | | 公開日 | 2016-10-05 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structural basis for disruption of claudin assembly in tight junctions by an enterotoxin
Sci Rep, 6, 2016
|
|
