3RP8
 
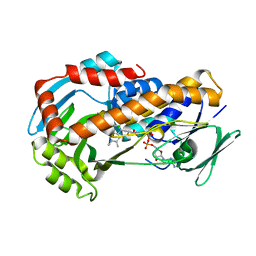 | | Crystal Structure of Klebsiella pneumoniae R204Q HpxO complexed with FAD | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, flavoprotein monooxygenase | | Authors: | Hicks, K.A, O'Leary, S.E, Begley, T.P, Ealick, S.E. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | Structural and Mechanistic Studies of HpxO, a Novel Flavin Adenine Dinucleotide-Dependent Urate Oxidase from Klebsiella pneumoniae.
Biochemistry, 52, 2013
|
|
3RP6
 
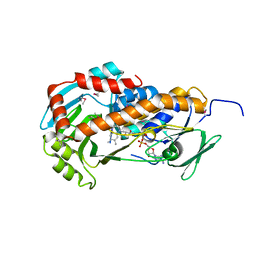 | | Crystal Structure of Klebsiella pneumoniae HpxO complexed with FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, flavoprotein monooxygenase | | Authors: | Hicks, K.A, O'Leary, S.E, Begley, T.P, Ealick, S.E. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Mechanistic Studies of HpxO, a Novel Flavin Adenine Dinucleotide-Dependent Urate Oxidase from Klebsiella pneumoniae.
Biochemistry, 52, 2013
|
|
3RP7
 
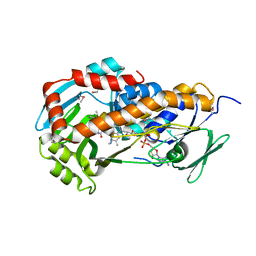 | | Crystal Structure of Klebsiella pneumoniae HpxO complexed with FAD and uric acid | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, URIC ACID, flavoprotein monooxygenase | | Authors: | Hicks, K.A, O'Leary, S.E, Begley, T.P, Ealick, S.E. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.042 Å) | | Cite: | Structural and Mechanistic Studies of HpxO, a Novel Flavin Adenine Dinucleotide-Dependent Urate Oxidase from Klebsiella pneumoniae.
Biochemistry, 52, 2013
|
|
3KVV
 
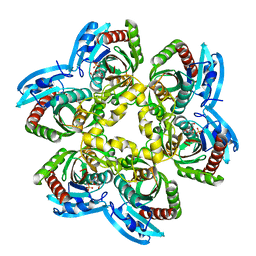 | | Trapping of an oxocarbenium ion intermediate in UP crystals | | Descriptor: | 1,4-anhydro-D-erythro-pent-1-enitol, 5-FLUOROURACIL, SULFATE ION, ... | | Authors: | Paul, D, O'Leary, S, Rajashankar, K, Bu, W, Toms, A, Settembre, E, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-11-30 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glycal formation in crystals of uridine phosphorylase.
Biochemistry, 49, 2010
|
|
3KVR
 
 | | Trapping of an oxocarbenium ion intermediate in UP crystals | | Descriptor: | 2,5-anhydro-4-deoxy-D-erythro-pent-4-enitol, 5-FLUOROURACIL, SULFATE ION, ... | | Authors: | Paul, D, O'Leary, S, Rajashankar, K, Bu, W, Toms, A, Settembre, E, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-11-30 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Glycal formation in crystals of uridine phosphorylase.
Biochemistry, 49, 2010
|
|
3KU4
 
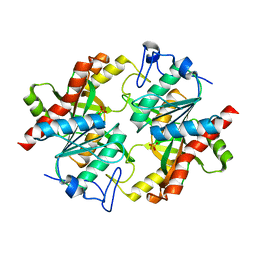 | | Trapping of an oxocarbenium ion intermediate in UP crystals | | Descriptor: | SULFATE ION, Uridine phosphorylase | | Authors: | Paul, D, O'Leary, S, Rajashankar, K, Bu, W, Toms, A, Settembre, E, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-11-26 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Glycal formation in crystals of uridine phosphorylase.
Biochemistry, 49, 2010
|
|
3KUK
 
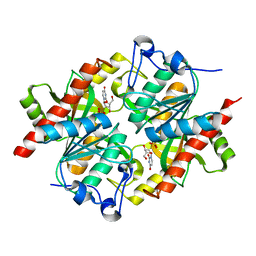 | | Trapping of an oxocarbenium ion intermediate in UP crystals | | Descriptor: | 2'-DEOXYURIDINE, SULFATE ION, Uridine phosphorylase | | Authors: | Paul, D, O'Leary, S, Rajashankar, K, Bu, W, Toms, A, Settembre, E, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-11-27 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.783 Å) | | Cite: | Glycal formation in crystals of uridine phosphorylase.
Biochemistry, 49, 2010
|
|
3KVY
 
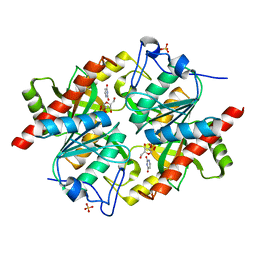 | | Trapping of an oxocarbenium ion intermediate in UP crystals | | Descriptor: | 1,4-anhydro-D-erythro-pent-1-enitol, SULFATE ION, URACIL, ... | | Authors: | Paul, D, O'Leary, S, Rajashankar, K, Bu, W, Toms, A, Settembre, E, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-11-30 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Glycal formation in crystals of uridine phosphorylase.
Biochemistry, 49, 2010
|
|
6WCZ
 
 | | CryoEM structure of full-length ZIKV NS5-hSTAT2 complex | | Descriptor: | Non-structural protein 5, Signal transducer and activator of transcription 2, ZINC ION | | Authors: | Boxiao, W, Stephanie, T, Kang, Z, Maria, T.S, Jian, F, Jiuwei, L, Linfeng, G, Wendan, R, Yanxiang, C, Ethan, C.V, HeaJin, H, Matthew, J.E, Sean, E.O, Adolfo, G.S, Hong, Z, Rong, H, Jikui, S. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7LMK
 
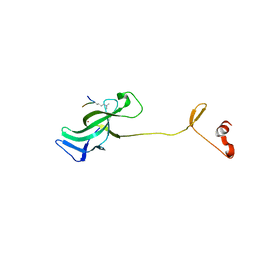 | |
7LMM
 
 | |
6UX2
 
 | | Crystal structure of ZIKV RdRp in complex with STAT2 | | Descriptor: | Nonstructural Protein 5, SULFATE ION, Signal transducer and activator of transcription 2, ... | | Authors: | Wang, B, Song, J. | | Deposit date: | 2019-11-06 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4X66
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Chen, J, Choi, J, Soltis, M, Puglisi, J.D. | | Deposit date: | 2014-12-06 | | Release date: | 2015-11-18 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (3.446 Å) | | Cite: | N(6)-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4X64
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Chen, J, Choi, J, Soltis, M, Puglisi, J.D. | | Deposit date: | 2014-12-06 | | Release date: | 2015-11-18 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | N(6)-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4X62
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Chen, J, Choi, J, Soltis, M, Puglisi, J.D. | | Deposit date: | 2014-12-06 | | Release date: | 2015-11-18 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (3.4492 Å) | | Cite: | N(6)-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4X65
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Chen, J, Choi, J, Soltis, M, Puglisi, J.D. | | Deposit date: | 2014-12-06 | | Release date: | 2015-11-18 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (3.345 Å) | | Cite: | N(6)-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics.
Nat.Struct.Mol.Biol., 23, 2016
|
|
6PZV
 
 | |
4JAT
 
 | |
4JAQ
 
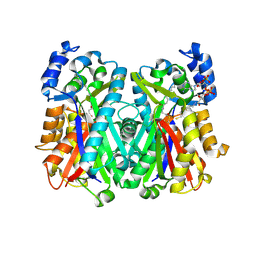 | | Crystal Structure of Mycobacterium tuberculosis PKS11 Reveals Intermediates in the Synthesis of Methyl-branched Alkylpyrones | | Descriptor: | 3,5-dioxoicosanoic acid, Alpha-pyrone synthesis polyketide synthase-like Pks11, COENZYME A, ... | | Authors: | Gokulan, K, Sacchettini, J.C, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2013-02-19 | | Release date: | 2013-04-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis polyketide synthase 11 (PKS11) reveals intermediates in the synthesis of methyl-branched alkylpyrones.
J.Biol.Chem., 288, 2013
|
|
4JD3
 
 | |
4JAP
 
 | |
4JAO
 
 | |
4JAR
 
 | |
