1RIO
 
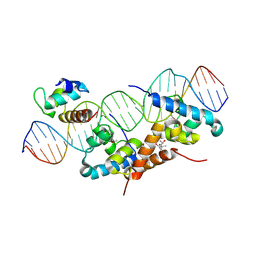 | | Structure of bacteriophage lambda cI-NTD in complex with sigma-region4 of Thermus aquaticus bound to DNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 27-MER, CALCIUM ION, ... | | Authors: | Jain, D, Nickels, B.E, Sun, L, Hochschild, A, Darst, S.A. | | Deposit date: | 2003-11-17 | | Release date: | 2004-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a ternary transcription activation complex.
Mol.Cell, 13, 2004
|
|
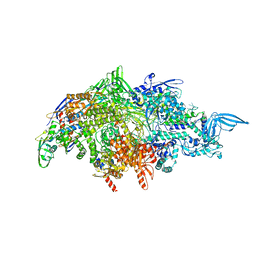
7RDQ
 
 | |
7EH0
 
 | |
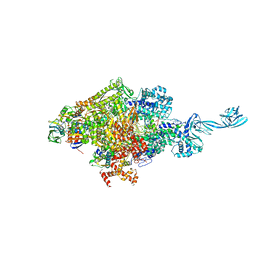
7EH2
 
 | |
7EH1
 
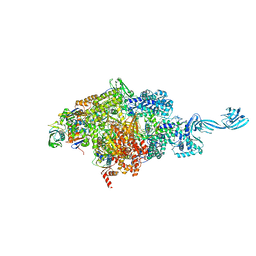 | | Thermus thermophilus transcription initiation complex containing a template-strand purine at position TSS-2, GpG RNA primer, and CMPcPP | | Descriptor: | 1,4-BUTANEDIOL, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, DNA (27-MER), ... | | Authors: | Li, L, Zhang, Y. | | Deposit date: | 2021-03-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Promoter-sequence determinants and structural basis of primer-dependent transcription initiation in Escherichia coli .
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2EYQ
 
 | |
3MLQ
 
 | |
5ULJ
 
 | |
5ULI
 
 | | Crystal Structure of mouse DXO in complex with (3'-NADP)+ and calcium ion | | Descriptor: | CALCIUM ION, Decapping and exoribonuclease protein, [[(2R,3S,4R,5R)-5-(3-aminocarbonyl-4H-pyridin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Doamekpor, S.K, Tong, L. | | Deposit date: | 2017-01-24 | | Release date: | 2017-05-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 5' End Nicotinamide Adenine Dinucleotide Cap in Human Cells Promotes RNA Decay through DXO-Mediated deNADding.
Cell, 168, 2017
|
|
7N4E
 
 | | Escherichia coli sigma 70-dependent paused transcription elongation complex | | Descriptor: | 5'-R(*UP*UP*CP*GP*GP*AP*GP*AP*GP*GP*UP*A)-3', DNA (61-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Molodtsov, V, Su, M, Ebright, R.H. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural and mechanistic basis of sigma-dependent transcriptional pausing.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7MLB
 
 | |
7MLI
 
 | |
7MLJ
 
 | |
7UBJ
 
 | | Transcription antitermination factor Qlambda, type-I crystal | | Descriptor: | 1,2-ETHANEDIOL, Antitermination protein Q, CHLORIDE ION, ... | | Authors: | Yin, Z, Ebright, R.H. | | Deposit date: | 2022-03-15 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | In transcription antitermination by Q lambda , NusA induces refolding of Q lambda to form a nozzle that extends the RNA polymerase RNA-exit channel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UBL
 
 | | Transcription antitermination factor Qlambda in complex with Q-lambda-binding-element DNA | | Descriptor: | 1,2-ETHANEDIOL, Antitermination protein Q, DNA (5'-D(P*CP*AP*CP*CP*CP*AP*AP*TP*TP*TP*TP*AP*TP*TP*CP*AP*AP*TP*G)-3'), ... | | Authors: | Yin, Z, Ebright, R.H. | | Deposit date: | 2022-03-15 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | In transcription antitermination by Q lambda , NusA induces refolding of Q lambda to form a nozzle that extends the RNA polymerase RNA-exit channel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UBM
 
 | |
7UBK
 
 | | Transcription antitermination factor Qlambda, type-II crystal | | Descriptor: | 1,2-ETHANEDIOL, Antitermination protein Q, CHLORIDE ION, ... | | Authors: | Yin, Z, Ebright, R.H. | | Deposit date: | 2022-03-15 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | In transcription antitermination by Q lambda , NusA induces refolding of Q lambda to form a nozzle that extends the RNA polymerase RNA-exit channel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UBN
 
 | |
5D4E
 
 | |
5E18
 
 | |
5E17
 
 | |
5D4C
 
 | |
5D4D
 
 | |
