4CYY
 
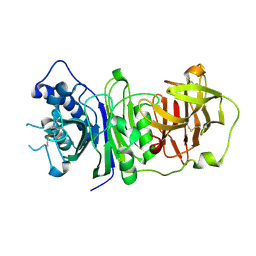 | | The structure of vanin-1: defining the link between metabolic disease, oxidative stress and inflammation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PANTETHEINASE | | Authors: | Boersma, Y.L, Newman, J, Adams, T.E, Sparrow, L, Cowieson, N, Lucent, D, Krippner, G, Bozaoglu, K, Peat, T.S. | | Deposit date: | 2014-04-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | The Structure of Vanin-1: A Key Enzyme Linking Metabolic Disease and Inflammation
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CYG
 
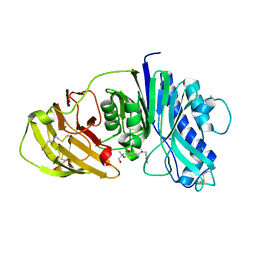 | | The structure of vanin-1: defining the link between metabolic disease, oxidative stress and inflammation | | Descriptor: | (2R)-2,4-dihydroxy-N-[(3S)-3-hydroxy-4-phenylbutyl]-3,3-dimethylbutanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Boersma, Y.L, Newman, J, Adams, T.E, Sparrow, L, Cowieson, N, Lucent, D, Krippner, G, Bozaoglu, K, Peat, T.S. | | Deposit date: | 2014-04-11 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of Vanin-1: A Key Enzyme Linking Metabolic Disease and Inflammation
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CYF
 
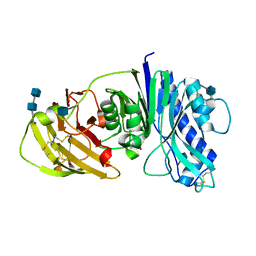 | | The structure of vanin-1: defining the link between metabolic disease, oxidative stress and inflammation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PANTETHEINASE | | Authors: | Boersma, Y.L, Newman, J, Adams, T.E, Sparrow, L, Cowieson, N, Lucent, D, Krippner, G, Bozaoglu, K, Peat, T.S. | | Deposit date: | 2014-04-11 | | Release date: | 2014-12-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Structure of Vanin-1: A Key Enzyme Linking Metabolic Disease and Inflammation
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CNW
 
 | | Surface residue engineering of bovine carbonic anhydrase to an extreme halophilic enzyme for potential application in postcombustion CO2 capture | | Descriptor: | CALCIUM ION, CARBONIC ANHYDRASE 2, ZINC ION | | Authors: | Warden, A, Newman, J, Peat, T.S, Seabrook, S, Williams, M, Dojchinov, G, Haritos, V. | | Deposit date: | 2014-01-25 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Rational Engineering of a Mesohalophilic Carbonic Anhydrase to an Extreme Halotolerant Biocatalyst.
Nat.Commun., 6, 2015
|
|
4CQC
 
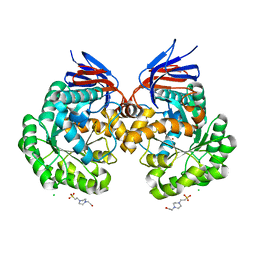 | | The reaction mechanism of the N-isopropylammelide isopropylaminohydrolase AtzC: insights from structural and mutagenesis studies | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, N-ISOPROPYLAMMELIDE ISOPROPYL AMIDOHYDROLASE, ... | | Authors: | Balotra, S, Newman, J, French, N.G, Peat, T.S, Scott, C. | | Deposit date: | 2014-02-13 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-Ray Structure and Mutagenesis Studies of the N-Isopropylammelide Isopropylaminohydrolase, Atzc
Plos One, 1, 2015
|
|
4CNX
 
 | | Surface residue engineering of bovine carbonic anhydrase to an extreme halophilic enzyme for potential application in postcombustion CO2 capture | | Descriptor: | CARBONIC ANHYDRASE 2, DI(HYDROXYETHYL)ETHER, ZINC ION | | Authors: | Warden, A, Newman, J, Peat, T.S, Seabrook, S, Williams, M, Dojchinov, G, Haritos, V. | | Deposit date: | 2014-01-25 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Rational Engineering of a Mesohalophilic Carbonic Anhydrase to an Extreme Halotolerant Biocatalyst.
Nat.Commun., 6, 2015
|
|
4CNV
 
 | | Surface residue engineering of bovine carbonic anhydrase to an extreme halophilic enzyme for potential application in postcombustion CO2 capture | | Descriptor: | CARBONIC ANHYDRASE 2, GLYCEROL, ZINC ION | | Authors: | Warden, A, Newman, J, Peat, T.S, Seabrook, S, Williams, M, Dojchinov, G, Haritos, V. | | Deposit date: | 2014-01-25 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Rational Engineering of a Mesohalophilic Carbonic Anhydrase to an Extreme Halotolerant Biocatalyst.
Nat.Commun., 6, 2015
|
|
4CNR
 
 | | Surface residue engineering of bovine carbonic anhydrase to an extreme halophilic enzyme for potential application in postcombustion CO2 capture | | Descriptor: | CARBONIC ANHYDRASE 2, ZINC ION | | Authors: | Warden, A, Newman, J, Peat, T.S, Seabrook, S, Williams, M, Dojchinov, G, Haritos, V. | | Deposit date: | 2014-01-24 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Rational Engineering of a Mesohalophilic Carbonic Anhydrase to an Extreme Halotolerant Biocatalyst.
Nat.Commun., 6, 2015
|
|
4CP8
 
 | | Structure of the amidase domain of allophanate hydrolase from Pseudomonas sp strain ADP | | Descriptor: | ALLOPHANATE HYDROLASE, MALONATE ION | | Authors: | Balotra, S, Newman, J, French, N, French, L, Peat, T.S, Scott, C. | | Deposit date: | 2014-02-03 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray Structure of the Amidase Domain of Atzf, the Allophanate Hydrolase from the Cyanuric Acid-Mineralizing Multienzyme Complex.
Appl.Environ.Microbiol., 81, 2015
|
|
4CQD
 
 | | The reaction mechanism of the N-isopropylammelide isopropylaminohydrolase AtzC: insights from structural and mutagenesis studies | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MALONATE ION, ... | | Authors: | Balotra, S, Newman, J, French, N.G, Peat, T.S, Scott, C. | | Deposit date: | 2014-02-13 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-Ray Structure and Mutagenesis Studies of the N-Isopropylammelide Isopropylaminohydrolase, Atzc
Plos One, 1, 2015
|
|
4CQB
 
 | | The reaction mechanism of the N-isopropylammelide isopropylaminohydrolase AtzC: insights from structural and mutagenesis studies | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, MALONATE ION, ... | | Authors: | Balotra, S, Newman, J, French, N.G, Peat, T.S, Scott, C. | | Deposit date: | 2014-02-13 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | X-Ray Structure and Mutagenesis Studies of the N-Isopropylammelide Isopropylaminohydrolase, Atzc
Plos One, 1, 2015
|
|
4B2G
 
 | | Crystal Structure of an Indole-3-Acetic Acid Amido Synthase from Vitis vinifera Involved in Auxin Homeostasis | | Descriptor: | GH3-1 AUXIN CONJUGATING ENZYME, MALONATE ION, [(2S,3R,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl] 2-(1H-indol-3-yl)ethyl hydrogen phosphate | | Authors: | Peat, T.S, Bottcher, C, Newman, J, Lucent, D, Cowieson, N, Davies, C. | | Deposit date: | 2012-07-16 | | Release date: | 2012-12-19 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of an Indole-3-Acetic Acid Amido Synthetase from Grapevine Involved in Auxin Homeostasis.
Plant Cell, 24, 2012
|
|
4BVR
 
 | | Cyanuric acid hydrolase: evolutionary innovation by structural concatenation. | | Descriptor: | 1,3,5-triazine-2,4,6-triol, CYANURIC ACID AMIDOHYDROLASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Peat, T.S, Balotra, S, Wilding, M, French, N.G, Briggs, L.J, Panjikar, S, Cowieson, N, Newman, J, Scott, C. | | Deposit date: | 2013-06-28 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Cyanuric Acid Hydrolase: Evolutionary Innovation by Structural Concatenation.
Mol.Microbiol., 88, 2013
|
|
4BVS
 
 | | Cyanuric acid hydrolase: evolutionary innovation by structural concatenation. | | Descriptor: | 1,3,5-triazine-2,4,6-triamine, CYANURIC ACID AMIDOHYDROLASE, MAGNESIUM ION | | Authors: | Peat, T.S, Balotra, S, Wilding, M, French, N.G, Briggs, L.J, Panjikar, S, Cowieson, N, Newman, J, Scott, C. | | Deposit date: | 2013-06-28 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cyanuric Acid Hydrolase: Evolutionary Innovation by Structural Concatenation.
Mol.Microbiol., 88, 2013
|
|
4BVT
 
 | | Cyanuric acid hydrolase: evolutionary innovation by structural concatenation | | Descriptor: | BARBITURIC ACID, CYANURIC ACID AMIDOHYDROLASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Peat, T.S, Balotra, S, Wilding, M, French, N.G, Briggs, L.J, Panjikar, S, Cowieson, N, Newman, J, Scott, C. | | Deposit date: | 2013-06-28 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Cyanuric Acid Hydrolase: Evolutionary Innovation by Structural Concatenation.
Mol.Microbiol., 88, 2013
|
|
4BVQ
 
 | | Cyanuric acid hydrolase: evolutionary innovation by structural concatenation. | | Descriptor: | CYANURIC ACID AMIDOHYDROLASE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Peat, T.S, Balotra, S, Wilding, M, French, N.G, Briggs, L.J, Panjikar, S, Cowieson, N, Newman, J, Scott, C. | | Deposit date: | 2013-06-28 | | Release date: | 2013-07-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cyanuric Acid Hydrolase: Evolutionary Innovation by Structural Concatenation.
Mol.Microbiol., 88, 2013
|
|
5TI8
 
 | |
6MAJ
 
 | | HBO1 is required for the maintenance of leukaemia stem cells | | Descriptor: | 4-fluoro-N'-[(3-hydroxyphenyl)sulfonyl]-5-methyl[1,1'-biphenyl]-3-carbohydrazide, BRD1 protein, GLYCEROL, ... | | Authors: | Ren, B, Peat, T.S, Monahan, B, Dawson, M, Street, I. | | Deposit date: | 2018-08-27 | | Release date: | 2019-12-25 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (2.139 Å) | | Cite: | HBO1 is required for the maintenance of leukaemia stem cells.
Nature, 577, 2020
|
|
6MAK
 
 | | HBO1 is required for the maintenance of leukaemia stem cells | | Descriptor: | ACETYL COENZYME *A, BRD1 protein, Histone acetyltransferase KAT7, ... | | Authors: | Ren, B, Peat, T.S, Monahan, B, Dawson, M, Street, I. | | Deposit date: | 2018-08-27 | | Release date: | 2019-12-25 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | HBO1 is required for the maintenance of leukaemia stem cells.
Nature, 577, 2020
|
|
3QBC
 
 | |
3PN1
 
 | | Novel Bacterial NAD+-dependent DNA Ligase Inhibitors with Broad Spectrum Potency and Antibacterial Efficacy In Vivo | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 2-(butylsulfanyl)adenosine, DNA ligase | | Authors: | Mills, S, Eakin, A, Buurman, E, Newman, J, Gao, N, Huynh, H, Johnson, K, Lahiri, S, Shapiro, A, Walkup, G, Wei, Y, Stokes, S. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel Bacterial NAD+-Dependent DNA Ligase Inhibitors with Broad-Spectrum Activity and Antibacterial Efficacy In Vivo.
Antimicrob.Agents Chemother., 55, 2011
|
|
6XA9
 
 | | SARS CoV-2 PLpro in complex with ISG15 C-terminal domain propargylamide | | Descriptor: | GLYCEROL, ISG15 CTD-propargylamide, Non-structural protein 3, ... | | Authors: | Klemm, T, Calleja, D.J, Richardson, L.W, Lechtenberg, B.C, Komander, D. | | Deposit date: | 2020-06-04 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2.
Embo J., 39, 2020
|
|
6XAA
 
 | | SARS CoV-2 PLpro in complex with ubiquitin propargylamide | | Descriptor: | Non-structural protein 3, Ubiquitin-propargylamide, ZINC ION | | Authors: | Klemm, T, Calleja, D.J, Richardson, L.W, Lechtenberg, B.C, Komander, D. | | Deposit date: | 2020-06-04 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2.
Embo J., 39, 2020
|
|
7PSP
 
 | | Crystal structure of S100A4 labeled with NU000846b. | | Descriptor: | (2R,4R)-1-(2-chloranylethanoyl)-N-(3-chlorophenyl)-4-phenyl-pyrrolidine-2-carboxamide, CALCIUM ION, Protein S100-A4 | | Authors: | Giroud, C, Szommer, T, Coxon, C, Monteiro, O, Christott, T, Bennett, J, Aitmakhanova, K, Raux, B, Newman, J, Elkins, J, Arruda Bezerra, G, Krojer, T, Koekemoer, L, Von Delft, F, Bountr, C, Brennan, P, Fedorov, O. | | Deposit date: | 2021-09-23 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of S100A4 labeled with NU000846b.
To Be Published
|
|
1IZ7
 
 | |
