3VKS
 
 | | Assimilatory nitrite reductase (Nii3) - NO complex from tobbaco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, NITRIC OXIDE, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-11-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The reductive reaction mechanism of tobacco nitrite reductase derived from a combination of crystal structures and ultraviolet-visible microspectroscopy
Proteins, 80, 2012
|
|
3VLY
 
 | | Assimilatory nitrite reductase (Nii3) - N226K mutant - SO3 partial complex from tobacco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Nitrite reductase, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray crystal structure of a mutant assimilatory nitrite reductase that shows sulfite reductase-like activity
Chem.Biodivers., 9, 2012
|
|
3VM0
 
 | | Assimilatory nitrite reductase (Nii3) - N226K mutant - NO2 complex from tobacco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, NITRITE ION, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | X-ray crystal structure of a mutant assimilatory nitrite reductase that shows sulfite reductase-like activity
Chem.Biodivers., 9, 2012
|
|
3VM1
 
 | | assimilatory nitrite reductase (Nii3) - N226K mutant - HCO3 complex from tobacco leaf | | Descriptor: | BICARBONATE ION, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystal structure of a mutant assimilatory nitrite reductase that shows sulfite reductase-like activity
Chem.Biodivers., 9, 2012
|
|
3VKP
 
 | | Assimilatory nitrite reductase (Nii3) - NO2 complex from tobbaco leaf analysed with low X-ray dose | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, NITRITE ION, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-11-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The reductive reaction mechanism of tobacco nitrite reductase derived from a combination of crystal structures and ultraviolet-visible microspectroscopy
Proteins, 80, 2012
|
|
3WWO
 
 | | S-selective hydroxynitrile lyase from Baliospermum montanum (apo1) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (S)-hydroxynitrile lyase, CALCIUM ION | | Authors: | Nakano, S, Dadashipour, M, Asano, Y. | | Deposit date: | 2014-06-23 | | Release date: | 2014-10-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and functional analysis of hydroxynitrile lyase from Baliospermum montanum with crystal structure, molecular dynamics and enzyme kinetics
Biochim.Biophys.Acta, 1844, 2014
|
|
3WWP
 
 | | S-selective hydroxynitrile lyase from Baliospermum montanum (apo2) | | Descriptor: | (S)-hydroxynitrile lyase, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Nakano, S, Dadashipour, M, Asano, Y. | | Deposit date: | 2014-06-23 | | Release date: | 2014-10-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of hydroxynitrile lyase from Baliospermum montanum with crystal structure, molecular dynamics and enzyme kinetics
Biochim.Biophys.Acta, 1844, 2014
|
|
6JNO
 
 | | RXRa structure complexed with CU-6PMN | | Descriptor: | 7-oxidanyl-2-oxidanylidene-6-(3,5,5,8,8-pentamethyl-6,7-dihydronaphthalen-2-yl)chromene-3-carboxylic acid, Retinoic acid receptor RXR-alpha | | Authors: | Kawasaki, M, Nakano, S, Motoyama, T, Yamada, S, Watanabe, M, Takamura, Y, Fujihara, M, Tokiwa, H, Kakuta, H, Ito, S. | | Deposit date: | 2019-03-17 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Competitive Binding Assay with an Umbelliferone-Based Fluorescent Rexinoid for Retinoid X Receptor Ligand Screening.
J.Med.Chem., 62, 2019
|
|
6JNR
 
 | | RXRa structure complexed with CU-6PMN and SRC1 peptide. | | Descriptor: | 7-oxidanyl-2-oxidanylidene-6-(3,5,5,8,8-pentamethyl-6,7-dihydronaphthalen-2-yl)chromene-3-carboxylic acid, HIS-LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN, Retinoic acid receptor RXR-alpha | | Authors: | Kawasaki, M, Nakano, S, Motoyama, T, Yamada, S, Watanabe, M, Takamura, Y, Fujihara, M, Tokiwa, H, Kakuta, H, Ito, S. | | Deposit date: | 2019-03-18 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RXRa structure complexed with CU-6PMN and SRC1 peptide.
To Be Published
|
|
8JHE
 
 | | Hyper-thermostable ancestral L-amino acid oxidase 2 (HTAncLAAO2) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Hyper thermostable ancestral L-amino acid oxidase | | Authors: | Kawamura, Y, Ishida, C, Miyata, R, Miyata, A, Hayashi, S, Fujinami, D, Ito, S, Nakano, S. | | Deposit date: | 2023-05-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural and functional analysis of hyper-thermostable ancestral L-amino acid oxidase that can convert Trp derivatives to D-forms by chemoenzymatic reaction.
Commun Chem, 6, 2023
|
|
8J82
 
 | | GaHNL-12gen (artificial S-hydroxynitrile lyase generated by GAOptimizer) | | Descriptor: | S-hydroxynitrile lyase | | Authors: | Ozawa, H, Unno, I, Sekine, R, Ito, S, Nakano, S. | | Deposit date: | 2023-04-29 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Development of evolutionary algorithm-based protein redesign method
Cell Rep Phys Sci, 5, 2024
|
|
6L96
 
 | | Structure of PPARalpha-LBD/pemafibrate/SRC1 peptide | | Descriptor: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, Peroxisome proliferator-activated receptor alpha, SRC1 coactivator peptide | | Authors: | Kawasaki, M, Kambe, A, Yamamoto, Y, Arulmozhira, S, Ito, S, Nakagawa, Y, Tokiwa, H, Nakano, S, Shimano, H. | | Deposit date: | 2019-11-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Elucidation of Molecular Mechanism of a Selective PPAR alpha Modulator, Pemafibrate, through Combinational Approaches of X-ray Crystallography, Thermodynamic Analysis, and First-Principle Calculations.
Int J Mol Sci, 21, 2020
|
|
4YK7
 
 | |
5ZQU
 
 | | Crystal structure of tetrameric RXRalpha-LBD complexed with partial agonist CBt-PMN | | Descriptor: | 1-(3,5,5,8,8-pentamethyl-6,7-dihydronaphthalen-2-yl)benzotriazole-5-carboxylic acid, BROMIDE ION, Retinoic acid receptor RXR-alpha | | Authors: | Miyashita, Y, Numoto, N, Arulmozhiraja, S, Nakano, S, Matsuo, N, Shimizu, K, Kakuta, H, Ito, S, Ikura, T, Ito, N, Tokiwa, H. | | Deposit date: | 2018-04-20 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.60038781 Å) | | Cite: | Dual conformation of the ligand induces the partial agonistic activity of retinoid X receptor alpha (RXR alpha ).
FEBS Lett., 593, 2019
|
|
7CFO
 
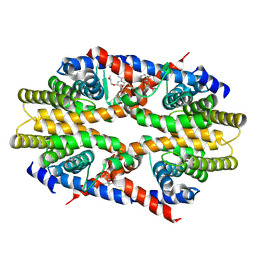 | | Crystal structure of human RXRalpha ligand binding domain complexed with CBTF-EE. | | Descriptor: | 1-[3-(2-ethoxyethoxy)-5,5,8,8-tetramethyl-6,7-dihydronaphthalen-2-yl]-2-(trifluoromethyl)benzimidazole-5-carboxylic acid, GLYCEROL, Retinoic acid receptor RXR-alpha | | Authors: | Watanabe, M, Fujihara, M, Motoyama, T, Kawasaki, M, Yamada, S, Takamura, Y, Ito, S, Makishima, M, Nakano, S, Kakuta, H. | | Deposit date: | 2020-06-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of a "Gatekeeper" Antagonist that Blocks Entry Pathway to Retinoid X Receptors (RXRs) without Allosteric Ligand Inhibition in Permissive RXR Heterodimers.
J.Med.Chem., 64, 2021
|
|
7BXS
 
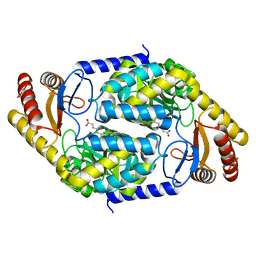 | | 2-amino-3-ketobutyrate CoA ligase from Cupriavidus necator glycine binding form | | Descriptor: | 2-amino-3-ketobutyrate coenzyme A ligase, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE] | | Authors: | Motoyama, T, Nakano, S, Hasebe, F, Miyoshi, N, Ito, S. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Chemoenzymatic synthesis of 3-ethyl-2,5-dimethylpyrazine by L-threonine 3-dehydrogenase and 2-amino-3-ketobutyrate CoA ligase/L-threonine aldolase
Commun Chem, 4, 2021
|
|
7BXQ
 
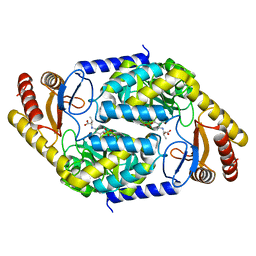 | | 2-amino-3-ketobutyrate CoA ligase from Cupriavidus necator L-Threonine binding form | | Descriptor: | 2-amino-3-ketobutyrate coenzyme A ligase, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-allothreonine | | Authors: | Motoyama, T, Nakano, S, Hasebe, F, Miyoshi, N, Ito, S. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Chemoenzymatic synthesis of 3-ethyl-2,5-dimethylpyrazine by L-threonine 3-dehydrogenase and 2-amino-3-ketobutyrate CoA ligase/L-threonine aldolase
Commun Chem, 4, 2021
|
|
7BXP
 
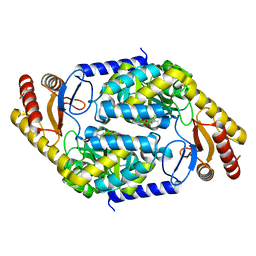 | | 2-amino-3-ketobutyrate CoA ligase from Cupriavidus necator | | Descriptor: | 2-amino-3-ketobutyrate coenzyme A ligase | | Authors: | Motoyama, T, Nakano, S, Hasebe, F, Miyoshi, N, Ito, S. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chemoenzymatic synthesis of 3-ethyl-2,5-dimethylpyrazine by L-threonine 3-dehydrogenase and 2-amino-3-ketobutyrate CoA ligase/L-threonine aldolase
Commun Chem, 4, 2021
|
|
7BXR
 
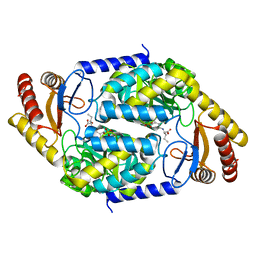 | | 2-amino-3-ketobutyrate CoA ligase from Cupriavidus necator 3-Hydroxynorvaline binding form | | Descriptor: | (2S,3R)-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-3-oxidanyl-pentanoic acid, 2-amino-3-ketobutyrate coenzyme A ligase | | Authors: | Motoyama, T, Nakano, S, Hasebe, F, Miyoshi, N, Ito, S. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Chemoenzymatic synthesis of 3-ethyl-2,5-dimethylpyrazine by L-threonine 3-dehydrogenase and 2-amino-3-ketobutyrate CoA ligase/L-threonine aldolase
Commun Chem, 4, 2021
|
|
7EIH
 
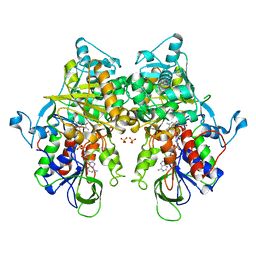 | | Ancestral L-Lys oxidase (ligand free form) | | Descriptor: | FAD dependent enzyme, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Sugiura, S, Nakano, S, Niwa, M, Hasebe, F, Ito, S. | | Deposit date: | 2021-03-31 | | Release date: | 2021-08-11 | | Last modified: | 2022-02-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic mechanism of ancestral L-lysine oxidase assigned by sequence data mining.
J.Biol.Chem., 297, 2021
|
|
7EII
 
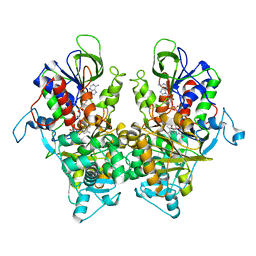 | | Ancestral L-Lys oxidase K387A variant (L-Lys binding form) | | Descriptor: | FAD dependent L-Lys oxidase, FLAVIN-ADENINE DINUCLEOTIDE, LYSINE | | Authors: | Sugiura, S, Nakano, S, Niwa, M, Hasebe, F, Ito, S. | | Deposit date: | 2021-03-31 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catalytic mechanism of ancestral L-lysine oxidase assigned by sequence data mining.
J.Biol.Chem., 297, 2021
|
|
7EIJ
 
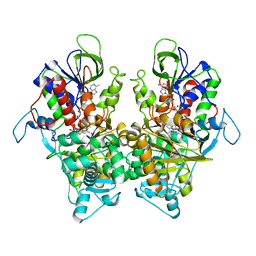 | | Ancestral L-Lys oxidase K387A variant (L-Arg binding form) | | Descriptor: | ARGININE, FAD dependent L-Lys oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Sugiura, S, Nakano, S, Niwa, M, Hasebe, F, Ito, S. | | Deposit date: | 2021-03-31 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic mechanism of ancestral L-lysine oxidase assigned by sequence data mining.
J.Biol.Chem., 297, 2021
|
|
5Y1F
 
 | | Monomeric L-threonine 3-dehydrogenase from metagenome database (NAD+ bound form) | | Descriptor: | NAD dependent epimerase/dehydratase family, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Motoyama, T, Nakano, S, Yamamoto, Y, Tokiwa, H, Asano, Y, Ito, S. | | Deposit date: | 2017-07-20 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Product Release Mechanism Associated with Structural Changes in Monomeric l-Threonine 3-Dehydrogenase.
Biochemistry, 56, 2017
|
|
5Y1G
 
 | | Monomeric L-threonine 3-dehydrogenase from metagenome database (AKB and NADH bound form) | | Descriptor: | 2-AMINO-3-KETOBUTYRIC ACID, NAD dependent epimerase/dehydratase family, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Motoyama, T, Nakano, S, Yamamoto, Y, Tokiwa, H, Asano, Y, Ito, S. | | Deposit date: | 2017-07-20 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Product Release Mechanism Associated with Structural Changes in Monomeric l-Threonine 3-Dehydrogenase.
Biochemistry, 56, 2017
|
|
5Y1E
 
 | | monomeric L-threonine 3-dehydrogenase from metagenome database (L-Ser and NAD+ bound form) | | Descriptor: | NAD dependent epimerase/dehydratase family, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SERINE | | Authors: | Motoyama, T, Nakano, S, Yamamoto, Y, Tokiwa, H, Asano, Y, Ito, S. | | Deposit date: | 2017-07-20 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Product Release Mechanism Associated with Structural Changes in Monomeric l-Threonine 3-Dehydrogenase.
Biochemistry, 56, 2017
|
|
