1TMN
 
 | |
1EE9
 
 | | CRYSTAL STRUCTURE OF THE NAD-DEPENDENT 5,10-METHYLENETETRAHYDROFOLATE DEHYDROGENASE FROM SACCHAROMYCES CEREVISIAE COMPLEXED WITH NAD | | Descriptor: | 5,10-METHYLENETETRAHYDROFOLATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Monzingo, A.F, Breksa, A, Ernst, S, Appling, D.R, Robertus, J.D. | | Deposit date: | 2000-01-31 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The X-ray structure of the NAD-dependent 5,10-methylenetetrahydrofolate dehydrogenase from Saccharomyces cerevisiae.
Protein Sci., 9, 2000
|
|
1EDZ
 
 | | STRUCTURE OF THE NAD-DEPENDENT 5,10-METHYLENETETRAHYDROFOLATE DEHYDROGENASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | 5,10-METHYLENETETRAHYDROFOLATE DEHYDROGENASE | | Authors: | Monzingo, A.F, Breksa, A, Ernst, S, Appling, D.R, Robertus, J.D. | | Deposit date: | 2000-01-28 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The X-ray structure of the NAD-dependent 5,10-methylenetetrahydrofolate dehydrogenase from Saccharomyces cerevisiae.
Protein Sci., 9, 2000
|
|
2IDR
 
 | | Crystal structure of translation initiation factor EIF4E from wheat | | Descriptor: | Eukaryotic translation initiation factor 4E-1 | | Authors: | Monzingo, A.F, Sadow, J, Dhaliwal, S, Lyon, A, Hoffman, D.W, Robertus, J.D, Browning, K.S. | | Deposit date: | 2006-09-15 | | Release date: | 2007-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of eukaryotic translation initiation factor-4E from wheat reveals a novel disulfide bond.
Plant Physiol., 143, 2007
|
|
2IDV
 
 | | Crystal structure of wheat C113S mutant EIF4E bound TO 7-methyl-GDP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Eukaryotic translation initiation factor 4E-1 | | Authors: | Monzingo, A.F, Dutt-Chaudhuri, A, Sadow, J, Dhaliwal, S, Hoffman, D.W, Robertus, J.D, Browning, K.S. | | Deposit date: | 2006-09-15 | | Release date: | 2007-06-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of eukaryotic translation initiation factor-4E from wheat reveals a novel disulfide bond.
Plant Physiol., 143, 2007
|
|
1PAF
 
 | | THE 2.5 ANGSTROMS STRUCTURE OF POKEWEED ANTIVIRAL PROTEIN | | Descriptor: | POKEWEED ANTIVIRAL PROTEIN | | Authors: | Monzingo, A.F, Collins, E.J, Ernst, S.R, Irvin, J.D, Robertus, J.D. | | Deposit date: | 1992-10-19 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 2.5 A structure of pokeweed antiviral protein.
J.Mol.Biol., 233, 1993
|
|
1PAG
 
 | | THE 2.5 ANGSTROMS STRUCTURE OF POKEWEED ANTIVIRAL PROTEIN | | Descriptor: | FORMYCIN-5'-MONOPHOSPHATE, POKEWEED ANTIVIRAL PROTEIN | | Authors: | Monzingo, A.F, Collins, E.J, Ernst, S.R, Irvin, J.D, Robertus, J.D. | | Deposit date: | 1992-10-19 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The 2.5 A structure of pokeweed antiviral protein.
J.Mol.Biol., 233, 1993
|
|
2NS6
 
 | | Crystal Structure of the Minimal Relaxase Domain of MobA from Plasmid R1162 | | Descriptor: | MANGANESE (II) ION, Mobilization protein A | | Authors: | Monzingo, A.F, Ozburn, A, Xia, S, Meyer, R.J, Robertus, J.D. | | Deposit date: | 2006-11-03 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of the Minimal Relaxase Domain of MobA at 2.1 A Resolution.
J.Mol.Biol., 366, 2007
|
|
1Q5X
 
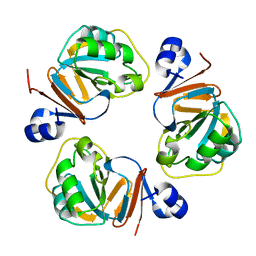 | | Structure of OF RRAA (MENG), a protein inhibitor of RNA processing | | Descriptor: | REGULATOR OF RNASE E ACTIVITY A | | Authors: | Monzingo, A.F, Gao, J, Qiu, J, Georgiou, G, Robertus, J.D. | | Deposit date: | 2003-08-11 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray Structure of Escherichia coli RraA (MenG), A Protein Inhibitor of RNA Processing.
J.Mol.Biol., 332, 2003
|
|
3RTJ
 
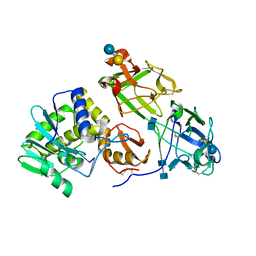 | | Crystal structure of ricin bound with dinucleotide ApG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, RNA (5'-R(*AP*G)-3'), Ricin A chain, ... | | Authors: | Monzingo, A.F, Robertus, J.D. | | Deposit date: | 2011-05-03 | | Release date: | 2011-08-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray analysis of substrate analogs in the ricin A-chain active site.
J.Mol.Biol., 227, 1992
|
|
3RTI
 
 | |
6OVZ
 
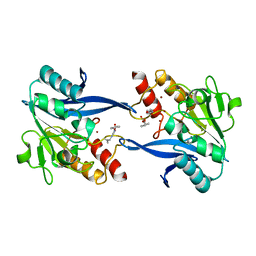 | | Crystal structure of the New Delhi metallo-beta-lactamase-1 adduct with a lysine-targeted affinity label | | Descriptor: | Beta-lactamase, CALCIUM ION, ZINC ION, ... | | Authors: | Monzingo, A.F, Fast, W, Thomas, P.W. | | Deposit date: | 2019-05-08 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.017 Å) | | Cite: | A Lysine-Targeted Affinity Label for Serine-beta-Lactamase Also Covalently Modifies New Delhi Metallo-beta-lactamase-1 (NDM-1).
Biochemistry, 58, 2019
|
|
3I2E
 
 | | Crystal structure of human dimethylarginine dymethylaminohydrolase-1 (DDAH-1) | | Descriptor: | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 | | Authors: | Monzingo, A.F, Wang, Y, Hu, S, Schaller, T.H, Robertus, J.D, Fast, W. | | Deposit date: | 2009-06-29 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Developing dual and specific inhibitors of dimethylarginine dimethylaminohydrolase-1 and nitric oxide synthase: toward a targeted polypharmacology to control nitric oxide.
Biochemistry, 48, 2009
|
|
3I4A
 
 | | Crystal structure of dimethylarginine dimethylaminohydrolase-1 (DDAH-1) in complex with N5-(1-iminopropyl)-L-ornithine | | Descriptor: | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1, N5-(1-iminopropyl)-L-ornithine | | Authors: | Monzingo, A.F, Wang, Y, Hu, S, Schaller, T.H, Fast, W, Robertus, J.D. | | Deposit date: | 2009-07-01 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Developing dual and specific inhibitors of dimethylarginine dimethylaminohydrolase-1 and nitric oxide synthase: toward a targeted polypharmacology to control nitric oxide.
Biochemistry, 48, 2009
|
|
6DGE
 
 | | Crystal structure of the dimethylarginine dimethylaminohydrolase adduct with N5-(1-imino-2-chloroethyl)-L-lysine | | Descriptor: | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1, N~6~-[(1E)-2-chloroethanimidoyl]-L-lysine | | Authors: | Monzingo, A.F, Burstein-Teitelbaum, G, Er, J.A.V, Tuley, A, Fast, W. | | Deposit date: | 2018-05-17 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Dissection, Optimization, and Structural Analysis of a Covalent Irreversible DDAH1 Inhibitor.
Biochemistry, 57, 2018
|
|
3BPB
 
 | | Crystal structure of the dimethylarginine dimethylaminohydrolase H162G adduct with S-methyl-L-thiocitrulline | | Descriptor: | N~5~-[(E)-imino(methylsulfanyl)methyl]-L-ornithine, dimethylarginine dimethylaminohydrolase | | Authors: | Monzingo, A.F, Linsky, T.W, Stone, E.M, Fast, W, Robertus, J.D. | | Deposit date: | 2007-12-18 | | Release date: | 2008-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Promiscuous partitioning of a covalent intermediate common in the pentein superfamily.
Chem.Biol., 15, 2008
|
|
3ETB
 
 | | Crystal structure of the engineered neutralizing antibody M18 complexed with anthrax protective antigen domain 4 | | Descriptor: | Anthrax Protective Antigen, Antibody M18 light chain and antibody M18 heavy chain linked with a synthetic (GGGGS)4 linker | | Authors: | Monzingo, A.F, Leysath, C.E, Barnett, J, Iverson, B.L, Georgiou, G, Robertus, J.D. | | Deposit date: | 2008-10-07 | | Release date: | 2009-04-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of the engineered neutralizing antibody M18 complexed to domain 4 of the anthrax protective antigen.
J.Mol.Biol., 387, 2009
|
|
3ESU
 
 | | Crystal structure of anthrax-neutralizing single-chain antibody 14b7 | | Descriptor: | Antibody 14b7* light chain and antibody 14b7* heavy chain linked with a synthetic (GGGGS)4 linker | | Authors: | Monzingo, A.F, Maynard, J.A, Iverson, B.L, Georgiou, G, Robertus, J.D. | | Deposit date: | 2008-10-06 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the engineered neutralizing antibody m18 complexed to domain 4 of the anthrax protective antigen.
J.Mol.Biol., 387, 2009
|
|
3ESV
 
 | | Crystal structure of the engineered neutralizing antibody M18 | | Descriptor: | Antibody M18 light chain and antibody M18 heavy chain linked with a synthetic (GGGGS)4 linker | | Authors: | Monzingo, A.F, Leysath, C.E, Barnett, J, Iverson, B.L, Georgiou, G, Robertus, J.D. | | Deposit date: | 2008-10-06 | | Release date: | 2009-04-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the engineered neutralizing antibody m18 complexed to domain 4 of the anthrax protective antigen.
J.Mol.Biol., 387, 2009
|
|
3RHY
 
 | | Crystal structure of the dimethylarginine dimethylaminohydrolase adduct with 4-chloro-2-hydroxymethylpyridine | | Descriptor: | (4-chloropyridin-2-yl)methanol, N(G),N(G)-dimethylarginine dimethylaminohydrolase | | Authors: | Monzingo, A.F, Johnson, C.M, Ke, Z, Yoon, D.-W, Linsky, T.W, Guo, H, Fast, W, Robertus, J.D. | | Deposit date: | 2011-04-12 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | On the mechanism of dimethylarginine dimethylaminohydrolase inactivation by 4-halopyridines.
J.Am.Chem.Soc., 133, 2011
|
|
3P8P
 
 | | Crystal Structure of Human Dimethylarginine Dimethylaminohydrolase-1 (DDAH-1) variant C274S bound with N5-(1-iminopentyl)-L-ornithine | | Descriptor: | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1, N~5~-[(1E)-pentanimidoyl]-L-ornithine | | Authors: | Monzingo, A.F, Lluis, M, Wang, Y, Fast, W, Robertus, J.D. | | Deposit date: | 2010-10-14 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterization of C-Alkyl Amidines as Bioavailable Covalent Reversible Inhibitors of Human DDAH-1.
Chemmedchem, 6, 2011
|
|
2TMN
 
 | | CRYSTALLOGRAPHIC STRUCTURAL ANALYSIS OF PHOSPHORAMIDATES AS INHIBITORS AND TRANSITION-STATE ANALOGS OF THERMOLYSIN | | Descriptor: | CALCIUM ION, N~2~-phosphono-L-leucinamide, Thermolysin, ... | | Authors: | Tronrud, D.E, Monzingo, A.F, Matthews, B.W. | | Deposit date: | 1987-06-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic structural analysis of phosphoramidates as inhibitors and transition-state analogs of thermolysin.
Eur.J.Biochem., 157, 1986
|
|
4TMN
 
 | | SLOW-AND FAST-BINDING INHIBITORS OF THERMOLYSIN DISPLAY DIFFERENT MODES OF BINDING. CRYSTALLOGRAPHIC ANALYSIS OF EXTENDED PHOSPHONAMIDATE TRANSITION-STATE ANALOGUES | | Descriptor: | CALCIUM ION, N-[(S)-[(1R)-1-{[(benzyloxy)carbonyl]amino}-2-phenylethyl](hydroxy)phosphoryl]-L-leucyl-L-alanine, THERMOLYSIN, ... | | Authors: | Holden, H.M, Tronrud, D.E, Monzingo, A.F, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1987-06-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Slow- and fast-binding inhibitors of thermolysin display different modes of binding: crystallographic analysis of extended phosphonamidate transition-state analogues.
Biochemistry, 26, 1987
|
|
5TMN
 
 | | Slow-and fast-binding inhibitors of thermolysin display different modes of binding. crystallographic analysis of extended phosphonamidate transition-state analogues | | Descriptor: | CALCIUM ION, N-[(S)-({[(benzyloxy)carbonyl]amino}methyl)(hydroxy)phosphoryl]-L-leucyl-L-leucine, THERMOLYSIN, ... | | Authors: | Holden, H.M, Tronrud, D.E, Monzingo, A.F, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1987-06-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Slow- and fast-binding inhibitors of thermolysin display different modes of binding: crystallographic analysis of extended phosphonamidate transition-state analogues.
Biochemistry, 26, 1987
|
|
2BAA
 
 | | THE REFINED CRYSTAL STRUCTURE OF AN ENDOCHITINASE FROM HORDEUM VULGARE L. SEEDS TO 1.8 ANGSTROMS RESOLUTION | | Descriptor: | ENDOCHITINASE (26 KD) | | Authors: | Hart, P.J, Pfluger, H.D, Monzingo, A.F, Ready, M.P, Ernst, S.R, Hollis, T, Robertus, J.D. | | Deposit date: | 1995-01-26 | | Release date: | 1996-01-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The refined crystal structure of an endochitinase from Hordeum vulgare L. seeds at 1.8 A resolution.
J.Mol.Biol., 248, 1995
|
|
