8USW
 
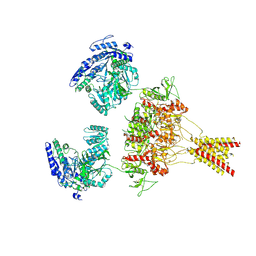 | | CNQX-bound GluN1a-3A NMDA receptor | | Descriptor: | 7-nitro-2,3-dioxo-1,2,3,4-tetrahydroquinoxaline-6-carbonitrile, Glutamate receptor ionotropic, NMDA 1, ... | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structure and function of GluN1-3A NMDA receptor excitatory glycine receptor channel.
Sci Adv, 10, 2024
|
|
8UUE
 
 | |
8USX
 
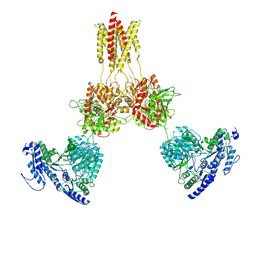 | | Glycine-bound GluN1a-3A NMDA receptor | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 3A | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-17 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure and function of GluN1-3A NMDA receptor excitatory glycine receptor channel.
Sci Adv, 10, 2024
|
|
8VUJ
 
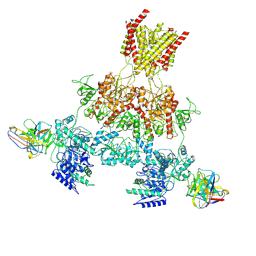 | | Human GluN1-2A with Fab 003-102 | | Descriptor: | 003-102 Heavy, 003-102 Light, Glutamate receptor ionotropic, ... | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-11 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structural and functional mechanisms of anti-NMDAR autoimmune encephalitis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8VVH
 
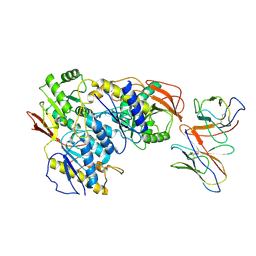 | | rat GluN1a-2B Fab 003-102 local refinement | | Descriptor: | 003-102 Heavy, 003-102 Light, Glutamate receptor ionotropic, ... | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2024-01-31 | | Release date: | 2024-09-11 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Structural and functional mechanisms of anti-NMDAR autoimmune encephalitis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8VUQ
 
 | |
8VUV
 
 | |
8VUT
 
 | | Human GluN1-2A with IgG 008-218 | | Descriptor: | 008-218 Heavy, 008-218 Light, Glutamate receptor ionotropic, ... | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-11 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural and functional mechanisms of anti-NMDAR autoimmune encephalitis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8VUH
 
 | | Human GluN1-2A IgG 003-102 splayed conformation | | Descriptor: | 003-102 Heavy, 003-102 Light, Glutamate receptor ionotropic, ... | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-11 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | Structural and functional mechanisms of anti-NMDAR autoimmune encephalitis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8VUL
 
 | |
8VUU
 
 | | Human GluN1-2B with Fab 007-168 | | Descriptor: | 007-168 Heavy, 007-168 Light, Glutamate receptor ionotropic, ... | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-11 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Structural and functional mechanisms of anti-NMDAR autoimmune encephalitis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8VUS
 
 | | Human GluN1-2A with IgG 007-168 | | Descriptor: | 007-168 Heavy, 007-168 Light, Glutamate receptor ionotropic, ... | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-11 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structural and functional mechanisms of anti-NMDAR autoimmune encephalitis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8VUR
 
 | | Human GluN1-2A with IgG 003-102 WT conformation | | Descriptor: | 003-102 Heavy, 003-102 Light, Glutamate receptor ionotropic, ... | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-11 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structural and functional mechanisms of anti-NMDAR autoimmune encephalitis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8VUN
 
 | | Human GluN1-2A With Fab 008-218 | | Descriptor: | 008-218 Heavy, 008-218 Light, Glutamate receptor ionotropic, ... | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-11 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Structural and functional mechanisms of anti-NMDAR autoimmune encephalitis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8VUY
 
 | | Rat GluN1-2B with Fab 003-102 | | Descriptor: | 003-102 Heavy, 003-102 Light, Glutamate receptor ionotropic, ... | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2024-01-30 | | Release date: | 2024-09-11 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structural and functional mechanisms of anti-NMDAR autoimmune encephalitis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6VD7
 
 | |
6VAL
 
 | |
6VAI
 
 | |
6VAM
 
 | |
6VAK
 
 | | Cryo-EM structure of human CALHM2 | | Descriptor: | Calcium homeostasis modulator protein 2 | | Authors: | Syrjanen, J.L, Chou, T.H, Furukawa, H. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structure and assembly of calcium homeostasis modulator proteins.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7SAB
 
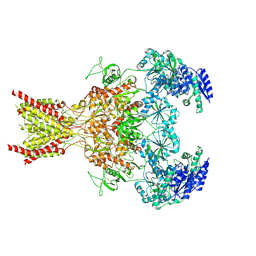 | | Phencyclidine-bound GluN1a-GluN2B NMDA receptors | | Descriptor: | 1-(PHENYL-1-CYCLOHEXYL)PIPERIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2021-09-22 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural insights into binding of therapeutic channel blockers in NMDA receptors.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SAD
 
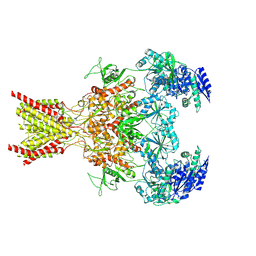 | | Memantine-bound GluN1a-GluN2B NMDA receptors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2021-09-22 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structural insights into binding of therapeutic channel blockers in NMDA receptors.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SAA
 
 | |
7SAC
 
 | | S-(+)-ketamine bound GluN1a-GluN2B NMDA receptors at 3.69 Angstrom resolution | | Descriptor: | (2~{S})-2-(2-chlorophenyl)-2-(methylamino)cyclohexan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2021-09-22 | | Release date: | 2022-07-20 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structural insights into binding of therapeutic channel blockers in NMDA receptors.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TES
 
 | |
