4YOI
 
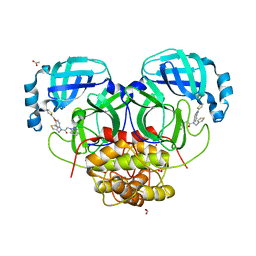 | | Structure of HKU4 3CLpro bound to non-covalent inhibitor 1A | | 分子名称: | 3C-like proteinase, ACETATE ION, FORMIC ACID, ... | | 著者 | St John, S.E, Mesecar, A. | | 登録日 | 2015-03-11 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Targeting zoonotic viruses: Structure-based inhibition of the 3C-like protease from bat coronavirus HKU4-The likely reservoir host to the human coronavirus that causes Middle East Respiratory Syndrome (MERS).
Bioorg.Med.Chem., 23, 2015
|
|
4YO9
 
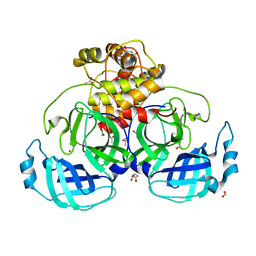 | | HKU4 3CLpro unbound structure | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3C-like proteinase, ACETATE ION, ... | | 著者 | St John, S.E, Mesecar, A. | | 登録日 | 2015-03-11 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Targeting zoonotic viruses: Structure-based inhibition of the 3C-like protease from bat coronavirus HKU4-The likely reservoir host to the human coronavirus that causes Middle East Respiratory Syndrome (MERS).
Bioorg.Med.Chem., 23, 2015
|
|
4YOJ
 
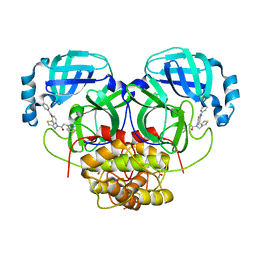 | | HKU4 3CLpro bound to non-covalent inhibitor 2A | | 分子名称: | 3C-like proteinase, ACETATE ION, FORMIC ACID, ... | | 著者 | St John, S.E, Mesecar, A. | | 登録日 | 2015-03-11 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Targeting zoonotic viruses: Structure-based inhibition of the 3C-like protease from bat coronavirus HKU4-The likely reservoir host to the human coronavirus that causes Middle East Respiratory Syndrome (MERS).
Bioorg.Med.Chem., 23, 2015
|
|
4YOG
 
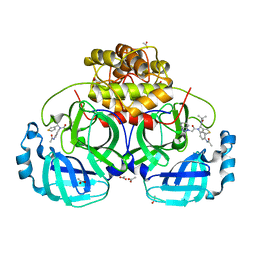 | | HKU4-3CLpro bound to non-covalent inhibitor 3B | | 分子名称: | 3C-like proteinase, ACETATE ION, N-[4-(acetylamino)phenyl]-2-(1H-benzotriazol-1-yl)-N-[(1R)-2-(tert-butylamino)-2-oxo-1-(thiophen-3-yl)ethyl]acetamide | | 著者 | St John, S.E, Mesecar, A. | | 登録日 | 2015-03-11 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Targeting zoonotic viruses: Structure-based inhibition of the 3C-like protease from bat coronavirus HKU4-The likely reservoir host to the human coronavirus that causes Middle East Respiratory Syndrome (MERS).
Bioorg.Med.Chem., 23, 2015
|
|
7JKV
 
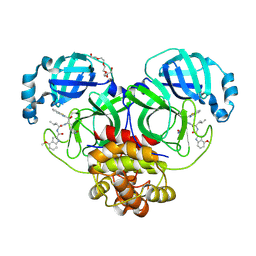 | | Crystal Structure of SARS-CoV-2 main protease in complex with an inhibitor GRL-2420 | | 分子名称: | 3C-like proteinase, N-[(2S)-1-({(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, PENTAETHYLENE GLYCOL | | 著者 | Bulut, H, Hattori, S.I, Das, D, Murayama, K, Mitsuya, H. | | 登録日 | 2020-07-29 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | A small molecule compound with an indole moiety inhibits the main protease of SARS-CoV-2 and blocks virus replication.
Nat Commun, 12, 2021
|
|
5V0N
 
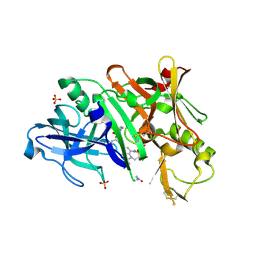 | | BACE1 in complex with inhibitor 5g | | 分子名称: | Beta-secretase 1, GLYCEROL, N-{(1S,2S)-1-[(2S)-4-benzyl-3-oxopiperazin-2-yl]-1-hydroxy-3-phenylpropan-2-yl}-7-ethyl-1,3,3-trimethyl-2,2-dioxo-1,2,3,4-tetrahydro-2lambda~6~-[1,2,5]thiadiazepino[3,4,5-hi]indole-9-carboxamide, ... | | 著者 | Mesecar, A, Ghosh, A, Yen, Y.-C. | | 登録日 | 2017-02-28 | | 公開日 | 2017-05-03 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.155 Å) | | 主引用文献 | Design, synthesis, and X-ray structural studies of BACE-1 inhibitors containing substituted 2-oxopiperazines as P1'-P2' ligands.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
7JYY
 
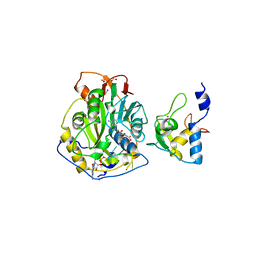 | | Crystal Structure of SARS-CoV-2 Nsp16/10 Heterodimer in Complex with (m7GpppA)pUpUpApApA (Cap-0) and S-Adenosylmethionine (SAM). | | 分子名称: | 2'-O-methyltransferase, CHLORIDE ION, FORMIC ACID, ... | | 著者 | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-09-01 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Mn 2+ coordinates Cap-0-RNA to align substrates for efficient 2'- O -methyl transfer by SARS-CoV-2 nsp16.
Sci.Signal., 14, 2021
|
|
1BL5
 
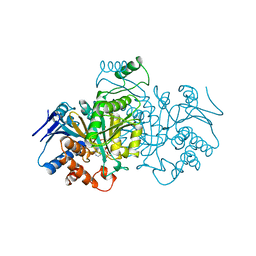 | | ISOCITRATE DEHYDROGENASE FROM E. COLI SINGLE TURNOVER LAUE STRUCTURE OF RATE-LIMITED PRODUCT COMPLEX, 10 MSEC TIME RESOLUTION | | 分子名称: | 2-OXOGLUTARIC ACID, ISOCITRATE DEHYDROGENASE, MAGNESIUM ION, ... | | 著者 | Stoddard, B.L, Cohen, B, Brubaker, M, Mesecar, A, Koshland Junior, D.E. | | 登録日 | 1998-07-23 | | 公開日 | 1999-05-04 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Millisecond Laue structures of an enzyme-product complex using photocaged substrate analogs.
Nat.Struct.Biol., 5, 1998
|
|
5DQC
 
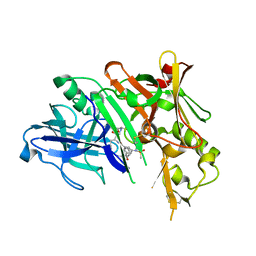 | | Co-crystal of BACE1 with compound 0211 | | 分子名称: | Beta-secretase 1, N-[(2S,3R)-3-hydroxy-4-({(2S,3S)-3-hydroxy-1-[(2-methylpropyl)amino]-1-oxobutan-2-yl}amino)-1-phenylbutan-2-yl]-5-[methyl(methylsulfonyl)amino]-N'-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide | | 著者 | Ghosh, A.K, Bhavanam, S.R, Yen, T.-C, Cardenas, E.L, Rao, K.V, Downs, D, Huang, X, Tang, J, Mescar, A.D. | | 登録日 | 2015-09-14 | | 公開日 | 2016-02-17 | | 最終更新日 | 2016-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.4651 Å) | | 主引用文献 | Design of Potent and Highly Selective Inhibitors for Human beta-Secretase 2 (Memapsin 1), a Target for Type 2 Diabetes.
Chem Sci, 7, 2016
|
|
4OW0
 
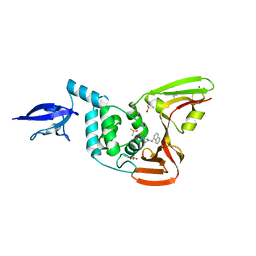 | |
4OVZ
 
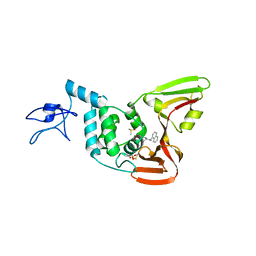 | |
2IDM
 
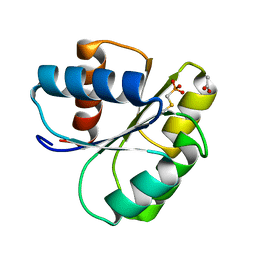 | | 2.00 A Structure of T87I/Y106W Phosphono-CheY | | 分子名称: | ACETATE ION, Chemotaxis protein cheY | | 著者 | Halkides, C.J, Haas, R.M, McAdams, K.A, Casper, E.S, Santarsiero, B.D, Mesecar, A.D. | | 登録日 | 2006-09-15 | | 公開日 | 2007-09-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The structures of T87I phosphono-CheY and T87I/Y106W phosphono-CheY help to explain their binding affinities to the FliM and CheZ peptides.
Arch.Biochem.Biophys., 479, 2008
|
|
2ID7
 
 | | 1.75 A Structure of T87I Phosphono-CheY | | 分子名称: | Chemotaxis protein cheY | | 著者 | Halkides, C.J, Haas, R.M, McAdams, K.A, Casper, E.S, Santarsiero, B.D, Mesecar, A.D. | | 登録日 | 2006-09-14 | | 公開日 | 2007-09-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The structures of T87I phosphono-CheY and T87I/Y106W phosphono-CheY help to explain their binding affinities to the FliM and CheZ peptides.
Arch.Biochem.Biophys., 479, 2008
|
|
2ID9
 
 | | 1.85 A Structure of T87I/Y106W Phosphono-CheY | | 分子名称: | Chemotaxis protein cheY | | 著者 | Halkides, C.J, Haas, R.M, McAdams, K.A, Casper, E.S, Santarsiero, B.D, Mesecar, A.D. | | 登録日 | 2006-09-14 | | 公開日 | 2007-09-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The structures of T87I phosphono-CheY and T87I/Y106W phosphono-CheY help to explain their binding affinities to the FliM and CheZ peptides.
Arch.Biochem.Biophys., 479, 2008
|
|
2DWU
 
 | | Crystal Structure of Glutamate Racemase Isoform RacE1 from Bacillus anthracis | | 分子名称: | D-GLUTAMIC ACID, GLYCEROL, Glutamate racemase, ... | | 著者 | Mehboob, S, Santarsiero, B.D, Johnson, M.E. | | 登録日 | 2006-08-17 | | 公開日 | 2007-06-19 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural and Functional Analysis of Two Glutamate Racemase Isozymes from Bacillus anthracis and Implications for Inhibitor Design
J.Mol.Biol., 371, 2007
|
|
4DEF
 
 | | Active site loop dynamics of a class IIa fructose 1,6-bisphosphate aldolase from M. tuberculosis | | 分子名称: | ACETATE ION, Fructose-bisphosphate aldolase, SODIUM ION, ... | | 著者 | Capodagli, G.C, Pegan, S.D. | | 登録日 | 2012-01-20 | | 公開日 | 2013-01-23 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Active site loop dynamics of a class IIa fructose 1,6-bisphosphate aldolase from Mycobacterium tuberculosis.
Biochemistry, 52, 2013
|
|
5TL6
 
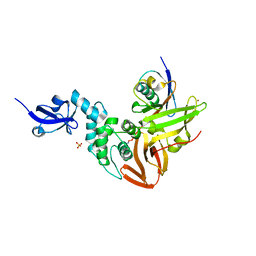 | | Crystal structure of SARS-CoV papain-like protease in complex with the C-terminal domain of human ISG15 | | 分子名称: | Replicase polyprotein 1ab, SULFATE ION, Ubiquitin-like protein ISG15, ... | | 著者 | Dzimianski, J.V, Daczkowski, C.M, Pegan, S.D. | | 登録日 | 2016-10-10 | | 公開日 | 2017-05-03 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.618 Å) | | 主引用文献 | Structural Insights into the Interaction of Coronavirus Papain-Like Proteases and Interferon-Stimulated Gene Product 15 from Different Species.
J. Mol. Biol., 429, 2017
|
|
5TLA
 
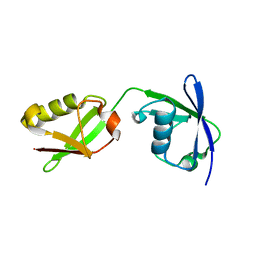 | |
5TL7
 
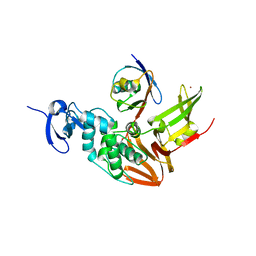 | |
3F4C
 
 | | Crystal structure of organophosphorus hydrolase from Geobacillus stearothermophilus strain 10, with glycerol bound | | 分子名称: | COBALT (II) ION, GLYCEROL, Organophosphorus hydrolase | | 著者 | Hawwa, R, Aikens, J, Turner, R.J, Santarsiero, B, Mesecar, A. | | 登録日 | 2008-10-31 | | 公開日 | 2009-09-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Structural basis for thermostability revealed through the identification and characterization of a highly thermostable phosphotriesterase-like lactonase from Geobacillus stearothermophilus.
Arch.Biochem.Biophys., 488, 2009
|
|
3F4D
 
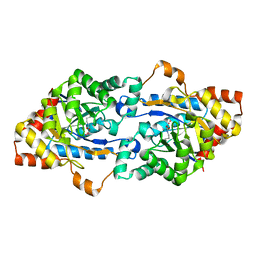 | | Crystal structure of organophosphorus hydrolase from Geobacillus stearothermophilus strain 10 | | 分子名称: | COBALT (II) ION, Organophosphorus hydrolase | | 著者 | Hawwa, R, Aikens, J, Turner, R.J, Santarsiero, B, Mesecar, A. | | 登録日 | 2008-10-31 | | 公開日 | 2009-08-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Structural basis for thermostability revealed through the identification and characterization of a highly thermostable phosphotriesterase-like lactonase from Geobacillus stearothermophilus.
Arch.Biochem.Biophys., 488, 2009
|
|
3GTX
 
 | | D71G/E101G mutant in organophosphorus hydrolase from Deinococcus radiodurans | | 分子名称: | COBALT (II) ION, Organophosphorus hydrolase | | 著者 | Hawwa, R, Larsen, S, Ratia, K, Mesecar, A. | | 登録日 | 2009-03-28 | | 公開日 | 2009-06-30 | | 最終更新日 | 2021-10-13 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Structure-based and random mutagenesis approaches increase the organophosphate-degrading activity of a phosphotriesterase homologue from Deinococcus radiodurans.
J.Mol.Biol., 393, 2009
|
|
3GTI
 
 | | D71G/E101G/M234L mutant in organophosphorus hydrolase from Deinococcus radiodurans | | 分子名称: | COBALT (II) ION, Organophosphorus hydrolase, SODIUM ION | | 著者 | Hawwa, R, Larsen, S, Ratia, K, Mesecar, A. | | 登録日 | 2009-03-27 | | 公開日 | 2009-06-30 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | Structure-based and random mutagenesis approaches increase the organophosphate-degrading activity of a phosphotriesterase homologue from Deinococcus radiodurans.
J.Mol.Biol., 393, 2009
|
|
3GU1
 
 | | Y97W mutant in organophosphorus hydrolase from Deinococcus radiodurans | | 分子名称: | COBALT (II) ION, GLYCEROL, Organophosphorus hydrolase | | 著者 | Hawwa, R, Larsen, S, Ratia, K, Mesecar, A. | | 登録日 | 2009-03-28 | | 公開日 | 2009-06-30 | | 最終更新日 | 2021-10-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-based and random mutagenesis approaches increase the organophosphate-degrading activity of a phosphotriesterase homologue from Deinococcus radiodurans.
J.Mol.Biol., 393, 2009
|
|
3GTH
 
 | | D71G/E101G/M234I mutant in organophosphorus hydrolase from Deinococcus radiodurans | | 分子名称: | COBALT (II) ION, FORMIC ACID, Organophosphorus hydrolase | | 著者 | Hawwa, R, Larsen, S, Ratia, K, Mesecar, A. | | 登録日 | 2009-03-27 | | 公開日 | 2009-06-30 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structure-based and random mutagenesis approaches increase the organophosphate-degrading activity of a phosphotriesterase homologue from Deinococcus radiodurans.
J.Mol.Biol., 393, 2009
|
|
