4NYV
 
 | | Crystal Structure of the Bromodomain of human CREBBP in complex with a quinazolin-one ligand | | Descriptor: | 6-bromo-3-methyl-3,4-dihydroquinazolin-2(1H)-one, CREB-binding protein | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Fedorov, O, Martin, S, Monteiro, O.P, von Delft, F, Brennan, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-11 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of the Bromodomain of human CREBBP in complex with a quinazolin-one ligand
TO BE PUBLISHED
|
|
4Q0N
 
 | | Crystal Structure of the fifth bromodomain of Human Poly-bromodomain containing protein 1 (PB1) in complex with a hydroxyphenyl-propenone ligand | | Descriptor: | (2E)-1-(2-hydroxyphenyl)-3-(2,4,5,7-tetrahydro-6H-pyrazolo[3,4-c]pyridin-6-yl)prop-2-en-1-one, 1,2-ETHANEDIOL, Protein polybromo-1 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Monteiro, O, Fedorov, O, Chaikuad, A, Yue, W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of the fifth bromodomain of Human Poly-bromodomain containing protein 1 (PB1) in complex with a hydroxyphenyl-propenone ligand
To be Published
|
|
4DMX
 
 | | Cathepsin K inhibitor | | Descriptor: | (1R,2R)-N-(1-cyanocyclopropyl)-2-{[4-(4-fluorophenyl)piperazin-1-yl]carbonyl}cyclohexanecarboxamide, Cathepsin K, GLYCEROL | | Authors: | Dossetter, A.G, Beeley, H, Bowyer, J, Cook, C.R, Crawford, J.J, Finlayson, J.E, Heron, N.M, Heyes, C, Highton, A.J, Hudson, J.A, Kenny, P.W, Martin, S, MacFaul, P.A, McGuire, T.M, Gutierrez, P.M, Morley, A.D, Morris, J.J, Page, K.M, Rosenbrier Ribeiro, L, Sawney, H, Steinbacher, S, Krapp, S, Jestel, A, Smith, C, Vickers, M. | | Deposit date: | 2012-02-08 | | Release date: | 2012-07-11 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | (1R,2R)-N-(1-cyanocyclopropyl)-2-(6-methoxy-1,3,4,5-tetrahydropyrido[4,3-b]indole-2-carbonyl)cyclohexanecarboxamide (AZD4996): a potent and highly selective cathepsin K inhibitor for the treatment of osteoarthritis.
J.Med.Chem., 55, 2012
|
|
4DMY
 
 | | Cathepsin K inhibitor | | Descriptor: | (1R,2R)-N-(1-cyanocyclopropyl)-2-[(8-fluoro-1,3,4,5-tetrahydro-2H-pyrido[4,3-b]indol-2-yl)carbonyl]cyclohexanecarboxamide, Cathepsin K, GLYCEROL, ... | | Authors: | Dossetter, A.G, Beeley, H, Bowyer, J, Cook, C.R, Crawford, J.J, Finlayson, J.E, Heron, N.M, Heyes, C, Highton, A.J, Hudson, J.A, Kenny, P.W, Martin, S, MacFaul, P.A, McGuire, T.M, Gutierrez, P.M, Morley, A.D, Morris, J.J, Page, K.M, Rosenbrier Ribeiro, L, Sawney, H, Steinbacher, S, Krapp, S, Jestel, A, Smith, C, Vickers, M. | | Deposit date: | 2012-02-08 | | Release date: | 2012-07-11 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | (1R,2R)-N-(1-cyanocyclopropyl)-2-(6-methoxy-1,3,4,5-tetrahydropyrido[4,3-b]indole-2-carbonyl)cyclohexanecarboxamide (AZD4996): a potent and highly selective cathepsin K inhibitor for the treatment of osteoarthritis.
J.Med.Chem., 55, 2012
|
|
8E1O
 
 | | Crystal structure of hTEAD2 bound to a methoxypyridine lipid pocket binder | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-methoxy-N-({3-[2-(methylamino)-2-oxoethyl]phenyl}methyl)-4-{(E)-2-[trans-4-(trifluoromethyl)cyclohexyl]ethenyl}pyridine-2-carboxamide, Transcriptional enhancer factor TEF-4 | | Authors: | Noland, C.L, Dey, A, Zbieg, J, Crawford, J. | | Deposit date: | 2022-08-10 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Targeting the Hippo pathway in cancers via ubiquitination dependent TEAD degradation
Biorxiv, 2024
|
|
6FKL
 
 | | Tubulin-TUB015 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-{1-[(2-Methoxyphenyl)amino]ethylidene}-5-phenyl-1,3-cyclohexanedione, CALCIUM ION, ... | | Authors: | Prota, A.E, Steinmetz, M.O, Priego, E.-M. | | Deposit date: | 2018-01-24 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | High-affinity ligands of the colchicine domain in tubulin based on a structure-guided design.
Sci Rep, 8, 2018
|
|
6FKJ
 
 | | Tubulin-TUB075 complex | | Descriptor: | (5~{S})-2-[(~{E})-~{N}-(2-ethoxyphenyl)-~{C}-methyl-carbonimidoyl]-3-oxidanyl-5-phenyl-cyclohex-2-en-1-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Prota, A.E, Steinmetz, M.O, Priego, E.-M. | | Deposit date: | 2018-01-24 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | High-affinity ligands of the colchicine domain in tubulin based on a structure-guided design.
Sci Rep, 8, 2018
|
|
2BZT
 
 | | NMR structure of the bacterial protein YFHJ from E. coli | | Descriptor: | PROTEIN ISCX | | Authors: | Pastore, C, Kelly, G, Adinolfi, S, Mc Cormick, J.E, Pastore, A. | | Deposit date: | 2005-08-22 | | Release date: | 2006-12-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | YfhJ, a molecular adaptor in iron-sulfur cluster formation or a frataxin-like protein?
Structure, 14, 2006
|
|
6QH5
 
 | | AP2 clathrin adaptor mu2T156-phosphorylated core in closed conformation | | Descriptor: | AP-2 complex subunit alpha, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Wrobel, A.G, Owen, D.J, McCoy, A.J, Evans, P.R. | | Deposit date: | 2019-01-15 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Dev.Cell, 50, 2019
|
|
6QH6
 
 | | AP2 clathrin adaptor core with two cargo peptides in open+ conformation | | Descriptor: | AP-2 complex subunit alpha, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Wrobel, A.G, Owen, D.J, McCoy, A.J, Evans, P.R. | | Deposit date: | 2019-01-15 | | Release date: | 2019-09-04 | | Last modified: | 2022-12-07 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Dev.Cell, 50, 2019
|
|
4J0R
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(R)-hydroxy(phenyl)methyl]phenol, Bromodomain-containing protein 4, ... | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, Hewings, D.S, von Delft, F, Conway, S.J, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-01-31 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Optimization of 3,5-dimethylisoxazole derivatives as potent bromodomain ligands.
J.Med.Chem., 56, 2013
|
|
6QH7
 
 | | AP2 clathrin adaptor mu2T156-phosphorylated core with two cargo peptides in open+ conformation | | Descriptor: | ADAPTOR-RELATED PROTEIN COMPLEX 2, MU 2 SUBUNIT, C-TERMINAL DOMAIN, ... | | Authors: | Wrobel, A.G, Owen, D.J, McCoy, A.J, Evans, P.R. | | Deposit date: | 2019-01-15 | | Release date: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Dev.Cell, 50, 2019
|
|
4J0S
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(S)-hydroxy(phenyl)methyl]phenol, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, Hewings, D.S, von Delft, F, Conway, S.J, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-01-31 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Optimization of 3,5-dimethylisoxazole derivatives as potent bromodomain ligands.
J.Med.Chem., 56, 2013
|
|
6RH6
 
 | | Solution structure and 1H, 13C and 15N chemical shift assignments for the complex of NECAP1 PHear domain with phosphorylated AP2 mu2 148-163 | | Descriptor: | AP-2 complex subunit mu, Adaptin ear-binding coat-associated protein 1 | | Authors: | Owen, D.J, Neuhaus, D, Yang, J.-C, Herrmann, T. | | Deposit date: | 2019-04-18 | | Release date: | 2019-09-04 | | Method: | SOLUTION NMR | | Cite: | Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Dev.Cell, 50, 2019
|
|
6RH5
 
 | | Solution structure and 1H, 13C and 15N chemical shift assignments for NECAP1 PHear domain | | Descriptor: | Adaptin ear-binding coat-associated protein 1 | | Authors: | Owen, D.J, Neuhaus, D, Yang, J.-C, Herrmann, T. | | Deposit date: | 2019-04-18 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Dev.Cell, 50, 2019
|
|
2OPU
 
 | |
5AK2
 
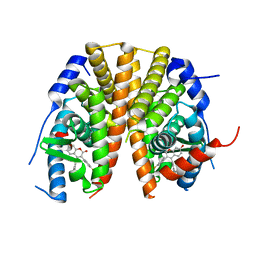 | | Oxyphenylpropenoic acids as Oral Selective Estrogen Receptor Down- Regulators. | | Descriptor: | (E)-3-[4-[[3-(4-fluoranyl-2-methyl-phenyl)-7-oxidanyl-2-oxidanylidene-chromen-4-yl]methyl]phenyl]prop-2-enoic acid, ESTROGEN RECEPTOR | | Authors: | Degorce, S, Bailey, A, Callis, R, De Savi, C, Ducray, R, Lamot, P, MacFaul, P, Maudet, M, Norman, R.A, Scott, J.S, Phillips, C. | | Deposit date: | 2015-02-27 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Investigation of (E)-3-[4-(2-Oxo-3-Aryl-Chromen-4-Yl)Oxyphenyl]Acrylic Acids as Oral Selective Estrogen Receptor Down-Regulators.
J.Med.Chem., 58, 2015
|
|
4NLD
 
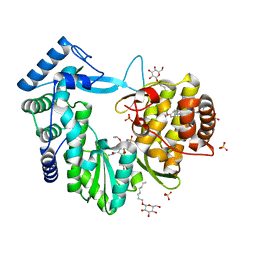 | | Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with BMS-791325 also known as (1aR,12bS)-8-cyclohexyl-n-(dimethylsulfamoyl)-11-methoxy-1a-{[(1R,5S)-3-methyl-3,8-diazabicyclo[3.2.1]oct-8-yl]carbonyl}-1,1a,2,12b-tetrahydrocyclopropa[d]indolo[2,1-a][2]benzazepine-5-carboxamide and 2-(4-fluorophenyl)-n-methyl-6-[(methylsulfonyl)amino]-5-(propan-2-yloxy)-1-benzofuran-3-carboxamide | | Descriptor: | (1aR,12bS)-8-cyclohexyl-N-(dimethylsulfamoyl)-11-methoxy-1a-{[(1R,5S)-3-methyl-3,8-diazabicyclo[3.2.1]oct-8-yl]carbonyl}-1,1a,2,12b-tetrahydrocyclopropa[d]indolo[2,1-a][2]benzazepine-5-carboxamide, 2-(4-fluorophenyl)-N-methyl-6-[(methylsulfonyl)amino]-5-(propan-2-yloxy)-1-benzofuran-3-carboxamide, RNA-directed RNA polymerase, ... | | Authors: | Sheriff, S. | | Deposit date: | 2013-11-14 | | Release date: | 2014-03-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery and Preclinical Characterization of the Cyclopropylindolobenzazepine BMS-791325, A Potent Allosteric Inhibitor of the Hepatitis C Virus NS5B Polymerase.
J.Med.Chem., 57, 2014
|
|
7TYU
 
 | | TEAD2 bound to Compound 2 | | Descriptor: | (3R)-1-[(8S)-5-(4-cyclohexylphenyl)-7-oxo-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonyl]pyrrolidine-3-carbonitrile, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Transcriptional enhancer factor TEF-4 | | Authors: | Noland, C.L, Fong, R. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Novel mechanism of YAP-TEAD inhibition results in targeted chromatin remodeling and reveals an
expanded Hippo dependent landscape in cancers
To Be Published
|
|
7TYP
 
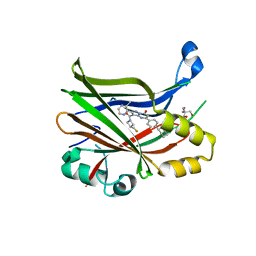 | | TEAD2 bound to GNE-7883 | | Descriptor: | (8S)-5-(4-cyclohexylphenyl)-3-[3-(fluoromethyl)azetidine-1-carbonyl]-2-(3-methylpyrazin-2-yl)pyrazolo[1,5-a]pyrimidin-7(4H)-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Transcriptional enhancer factor TEF-4 | | Authors: | Noland, C.L, Fong, R. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Novel mechanism of YAP-TEAD inhibition results in targeted chromatin remodeling and reveals an
expanded Hippo dependent landscape in cancers
To Be Published
|
|
7TYQ
 
 | | TEAD2 bound to Compound 1 | | Descriptor: | Transcriptional enhancer factor TEF-4, ethyl (8S)-7-oxo-5-[4-(trifluoromethyl)phenyl]-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carboxylate | | Authors: | Noland, C.L, Fong, R. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Novel mechanism of YAP-TEAD inhibition results in targeted chromatin remodeling and reveals an
expanded Hippo dependent landscape in cancers
To Be Published
|
|
4GPJ
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a isoxazolylbenzimidazole ligand | | Descriptor: | (1R)-6-(3,5-dimethyl-1,2-oxazol-4-yl)-1-phenyl-2,3-dihydro-1H-inden-1-ol, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, Heightman, T.D, Brennan, P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The design and synthesis of 5- and 6-isoxazolylbenzimidazoles as selective inhibitors of the BET bromodomains.
Medchemcomm, 4, 2013
|
|
3IIY
 
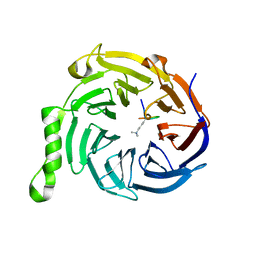 | | Crystal structure of Eed in complex with a trimethylated histone H1K26 peptide | | Descriptor: | Histone H1K26 peptide, Polycomb protein EED | | Authors: | Justin, N, Sharpe, M.L, Martin, S, Taylor, W.R, De Marco, V, Gamblin, S.J. | | Deposit date: | 2009-08-03 | | Release date: | 2009-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Role of the polycomb protein EED in the propagation of repressive histone marks.
Nature, 461, 2009
|
|
3IIW
 
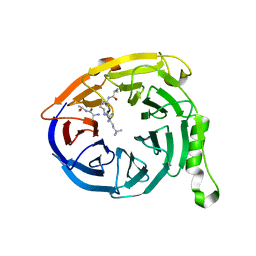 | | Crystal structure of Eed in complex with a trimethylated histone H3K27 peptide | | Descriptor: | Histone H3 peptide, Polycomb protein EED | | Authors: | Justin, N, Sharpe, M.L, Martin, S, Taylor, W.R, De Marco, V, Gamblin, S.J. | | Deposit date: | 2009-08-03 | | Release date: | 2009-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of the polycomb protein EED in the propagation of repressive histone marks.
Nature, 461, 2009
|
|
3IJ0
 
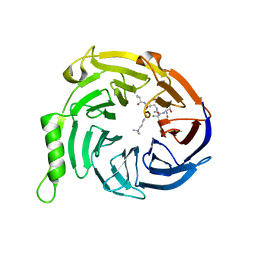 | | Crystal structure of Eed in complex with a trimethylated histone H3K9 peptide | | Descriptor: | Histone H3K9 peptide, Polycomb protein EED | | Authors: | Justin, N, Sharpe, M.L, Martin, S, Taylor, W.R, De Marco, V, Gamblin, S.J. | | Deposit date: | 2009-08-03 | | Release date: | 2009-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Role of the polycomb protein EED in the propagation of repressive histone marks.
Nature, 461, 2009
|
|
