5F6J
 
 | |
7LAA
 
 | |
7LAB
 
 | |
7KNX
 
 | | Crystal structure of SND1 in complex with C-26-A6 | | Descriptor: | 5-chloro-2-methoxy-N-(2-methyl[1,2,4]triazolo[1,5-a]pyridin-8-yl)benzene-1-sulfonamide, GLYCEROL, Staphylococcal nuclease domain-containing protein 1, ... | | Authors: | Kang, Y. | | Deposit date: | 2020-11-06 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Small-molecule inhibitors that disrupt the MTDH-SND1 complex suppress breast cancer progression and metastasis.
Nat Cancer, 3, 2022
|
|
7KNW
 
 | | Crystal structure of SND1 in complex with C-26-A2 | | Descriptor: | 5-chloro-2-methoxy-N-([1,2,4]triazolo[1,5-a]pyridin-8-yl)benzene-1-sulfonamide, GLYCEROL, Staphylococcal nuclease domain-containing protein 1 | | Authors: | Kang, Y. | | Deposit date: | 2020-11-06 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Small-molecule inhibitors that disrupt the MTDH-SND1 complex suppress breast cancer progression and metastasis.
Nat Cancer, 3, 2022
|
|
7LD1
 
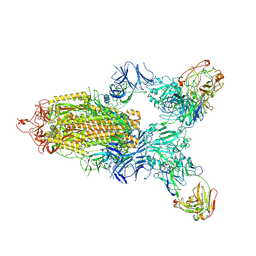 | |
7LCN
 
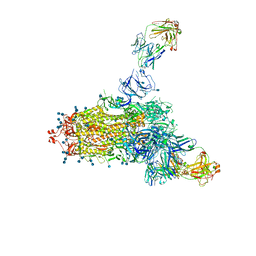 | |
6UM5
 
 | | Cryo-EM structure of HIV-1 neutralizing antibody DH270 UCA3 in complex with CH848 10.17DT Env | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Acharya, P, Henderson, R.C, Saunders, K.O, Haynes, B.F. | | Deposit date: | 2019-10-09 | | Release date: | 2019-12-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Targeted selection of HIV-specific antibody mutations by engineering B cell maturation.
Science, 366, 2019
|
|
6UM6
 
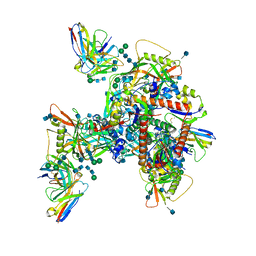 | | Cryo-EM structure of HIV-1 neutralizing antibody DH270.6 in complex with CH848 10.17DT Env | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Acharya, P, Henderson, R.C, Saunder, K.O, Haynes, B.F. | | Deposit date: | 2019-10-09 | | Release date: | 2019-12-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Targeted selection of HIV-specific antibody mutations by engineering B cell maturation.
Science, 366, 2019
|
|
6UM7
 
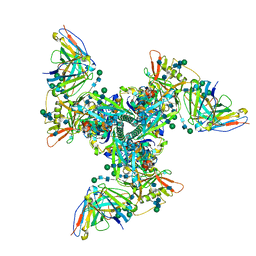 | | Cryo-EM structure of vaccine-elicited HIV-1 neutralizing antibody DH270.mu1 in complex with CH848 10.17DT Env | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH270.mu1 Fab Heavy Chain, DH270.mu1 Fab Light chain, ... | | Authors: | Acharya, P, Henderson, R.C, Saunders, K, Haynes, B.F. | | Deposit date: | 2019-10-09 | | Release date: | 2019-12-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Targeted selection of HIV-specific antibody mutations by engineering B cell maturation.
Science, 366, 2019
|
|
7X9E
 
 | | Crystal structure of the 76E1 Fab in complex with a SARS-CoV-2 spike peptide | | Descriptor: | 76E1 Fab Heavy Chain, 76E1 Fab Light Chain, Spike peptide | | Authors: | Chen, X, Zhang, T, Ding, J, Sun, X, Sun, B. | | Deposit date: | 2022-03-15 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Neutralization mechanism of a human antibody with pan-coronavirus reactivity including SARS-CoV-2.
Nat Microbiol, 7, 2022
|
|
4QUW
 
 | |
4RC7
 
 | |
4RC8
 
 | |
4RC5
 
 | |
6CTE
 
 | | 77Se-NMR probes the protein environment of selenomethionine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Chen, Q, Rozovsky, S. | | Deposit date: | 2018-03-22 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | 77Se NMR Probes the Protein Environment of Selenomethionine.
J.Phys.Chem.B, 124, 2020
|
|
6CPZ
 
 | | Selenomethionine mutant (I6Sem) of protein GB1 examined by X-ray diffraction | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Immunoglobulin G-binding protein G, ... | | Authors: | Chen, Q, Rozovsky, S. | | Deposit date: | 2018-03-14 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | 77Se NMR Probes the Protein Environment of Selenomethionine.
J.Phys.Chem.B, 124, 2020
|
|
6CHE
 
 | |
6CNE
 
 | | Selenomethionine variant (V29SeM) of protein GB1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Immunoglobulin G-binding protein G, PHOSPHATE ION | | Authors: | Chen, Q. | | Deposit date: | 2018-03-08 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | 77Se NMR Probes the Protein Environment of Selenomethionine.
J.Phys.Chem.B, 124, 2020
|
|
6GWR
 
 | | Structure of the kinase domain of human DDR1 in complex with a potent and selective inhibitor of DDR1 and DDR2 | | Descriptor: | 1,2-ETHANEDIOL, 3-(2-imidazo[1,2-a]pyrazin-3-ylethynyl)-~{N}-[3-[(4-methylpiperazin-1-yl)methyl]-5-(trifluoromethyl)phenyl]-4-propan-2-yl-benzamide, Epithelial discoidin domain-containing receptor 1, ... | | Authors: | Pinkas, D.M, Fox, A.E, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-06-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 3-(Imidazo[1,2- a]pyrazin-3-ylethynyl)-4-isopropyl- N-(3-((4-methylpiperazin-1-yl)methyl)-5-(trifluoromethyl)phenyl)benzamide as a Dual Inhibitor of Discoidin Domain Receptors 1 and 2.
J. Med. Chem., 61, 2018
|
|
8J5X
 
 | | The crystal structure of TrkA(G595R) kinase in complex with N-(3-cyclopropyl-5-((4-methylpiperazin-1-yl)methyl)phenyl)-4^6-methyl-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | Descriptor: | High affinity nerve growth factor receptor, N-(3-cyclopropyl-5-((4-methylpiperazin-1-yl)methyl)phenyl)-4^6-methyl-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | Authors: | Zhang, Z.M, Wang, Y.J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.09192252 Å) | | Cite: | Structure-Based Optimization of the Third Generation Type II Macrocycle TRK Inhibitors with Improved Activity against Solvent-Front, xDFG, and Gatekeeper Mutations.
J.Med.Chem., 66, 2023
|
|
8J61
 
 | | The crystal structure of TrkA kinase in complex with 4^6-methyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | Descriptor: | 4^6-methyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide, High affinity nerve growth factor receptor | | Authors: | Zhang, Z.M, Wang, Y.J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.05065274 Å) | | Cite: | Structure-Based Optimization of the Third Generation Type II Macrocycle TRK Inhibitors with Improved Activity against Solvent-Front, xDFG, and Gatekeeper Mutations.
J.Med.Chem., 66, 2023
|
|
8J63
 
 | | The crystal structure of TrkA kinase in complex with 4^6-methyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-11-oxo-5-oxa-10,14-diaza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | Descriptor: | 4^6-methyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-11-oxo-5-oxa-10,14-diaza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide, High affinity nerve growth factor receptor | | Authors: | Zhang, Z.M, Wang, Y.J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.0005 Å) | | Cite: | Structure-Based Optimization of the Third Generation Type II Macrocycle TRK Inhibitors with Improved Activity against Solvent-Front, xDFG, and Gatekeeper Mutations.
J.Med.Chem., 66, 2023
|
|
8J5W
 
 | | The crystal structure of TrkA(F589L) kinase in complex with N-(3-cyclopropyl-5-((4-methylpiperazin-1-yl)methyl)phenyl)-4^6-methyl-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | Descriptor: | High affinity nerve growth factor receptor, N-(3-cyclopropyl-5-((4-methylpiperazin-1-yl)methyl)phenyl)-4^6-methyl-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | Authors: | Zhang, Z.M, Wang, Y.J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.28041458 Å) | | Cite: | Structure-Based Optimization of the Third Generation Type II Macrocycle TRK Inhibitors with Improved Activity against Solvent-Front, xDFG, and Gatekeeper Mutations.
J.Med.Chem., 66, 2023
|
|
6KMH
 
 | | The crystal structure of CASK/Mint1 complex | | Descriptor: | Amyloid-beta A4 precursor protein-binding family A member 1, CHLORIDE ION, IODIDE ION, ... | | Authors: | Li, W, Feng, W. | | Deposit date: | 2019-07-31 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CASK modulates the assembly and function of the Mint1/Munc18-1 complex to regulate insulin secretion.
Cell Discov, 6, 2020
|
|
