1V1I
 
 | | Adenovirus fibre shaft sequence N-terminally fused to the bacteriophage T4 fibritin foldon trimerisation motif with a long linker | | Descriptor: | FIBRITIN, FIBER PROTEIN | | Authors: | Papanikolopoulou, K, Teixeira, S, Belrhali, H, Forsyth, V.T, Mitraki, A, van Raaij, M.J. | | Deposit date: | 2004-04-16 | | Release date: | 2004-07-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Adenovirus Fibre Shaft Sequences Fold Into the Native Triple Beta-Spiral Fold When N-Terminally Fused to the Bacteriophage T4 Fibritin Foldon Trimerisation Motif
J.Mol.Biol., 342, 2004
|
|
1V1H
 
 | | Adenovirus fibre shaft sequence N-terminally fused to the bacteriophage T4 fibritin foldon trimerisation motif with a short linker | | Descriptor: | FIBRITIN, FIBER PROTEIN | | Authors: | Papanikolopoulou, K, Teixeira, S, Belrhali, H, Forsyth, V.T, Mitraki, A, van Raaij, M.J. | | Deposit date: | 2004-04-16 | | Release date: | 2004-07-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Adenovirus Fibre Shaft Sequences Fold Into the Native Triple Beta-Spiral Fold When N-Terminally Fused to the Bacteriophage T4 Fibritin Foldon Trimerisation Motif
J.Mol.Biol., 342, 2004
|
|
4JN3
 
 | |
4JN5
 
 | |
5JI0
 
 | | PPARgamma-RXRalpha(S427F) heterodimer in complex with SRC-1, rosiglitazone, and 9-cis-retanoic acid | | Descriptor: | (9cis)-retinoic acid, 2,4-THIAZOLIDIINEDIONE, 5-[[4-[2-(METHYL-2-PYRIDINYLAMINO)ETHOXY]PHENYL]METHYL]-(9CL), ... | | Authors: | Bloudoff, K, Larsen, N.A. | | Deposit date: | 2016-04-21 | | Release date: | 2017-04-26 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | PPARgamma-RXRalpha(S427F) heterodimer in complex with SRC-1, rosiglitazone, and 9-cis-retanoic acid
To Be Published
|
|
3FEC
 
 | | Crystal structure of human Glutamate Carboxypeptidase III (GCPIII/NAALADase II), pseudo-unliganded | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Barinka, C, Lubkowski, J, Hlouchova, K. | | Deposit date: | 2008-11-28 | | Release date: | 2009-08-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural insight into the evolutionary and pharmacologic homology of glutamate carboxypeptidases II and III
Febs J., 276, 2009
|
|
3FEE
 
 | | The high resolution structure of human glutamate carboxypeptidase III (GCPIII/NAALADase II) in complex with quisqualic acid | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lubkowski, J, Barinka, C, Hlouchova, K. | | Deposit date: | 2008-11-28 | | Release date: | 2009-08-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural insight into the evolutionary and pharmacologic homology of glutamate carboxypeptidases II and III
Febs J., 276, 2009
|
|
3FED
 
 | | The high resolution structure of human glutamate carboxypeptidase III (GCPIII/NAALADase II) in complex with a transition state analog of Glu-Glu | | Descriptor: | (2S)-2-{[(S)-[(3S)-3-amino-3-carboxypropyl](hydroxy)phosphoryl]methyl}pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lubkowski, J, Barinka, C, Hlouchova, K. | | Deposit date: | 2008-11-28 | | Release date: | 2009-08-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structural insight into the evolutionary and pharmacologic homology of glutamate carboxypeptidases II and III
Febs J., 276, 2009
|
|
6P8F
 
 | | Crystal structure of CDK4 in complex with CyclinD1 and P27 | | Descriptor: | Cyclin-dependent kinase 4, Cyclin-dependent kinase inhibitor 1B, G1/S-specific cyclin-D1 | | Authors: | Guiley, K.Z, Stevenson, J.W, Lou, K, Barkovich, K.J, Bunch, K, Tripathi, S.M, Shokat, K.M, Rubin, S.M. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition.
Science, 366, 2019
|
|
6P8H
 
 | | Crystal structure of CDK4 in complex with CyclinD1 and P21 | | Descriptor: | Cyclin-dependent kinase 4, Cyclin-dependent kinase inhibitor 1, G1/S-specific cyclin-D1 | | Authors: | Guiley, K.Z, Stevenson, J.W, Lou, K, Barkovich, K.J, Bunch, K, Tripathi, S.M, Shokat, K.M, Rubin, S.M. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition.
Science, 366, 2019
|
|
6P8G
 
 | | Crystal structure of CDK4 in complex with CyclinD1 and P27 | | Descriptor: | Cyclin-dependent kinase 4, Cyclin-dependent kinase inhibitor 1B, G1/S-specific cyclin-D1 | | Authors: | Guiley, K.Z, Stevenson, J.W, Lou, K, Barkovich, K.J, Bunch, K, Tripathi, S.M, Shokat, K.M, Rubin, S.M. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition.
Science, 366, 2019
|
|
6P8E
 
 | | Crystal structure of CDK4 in complex with CyclinD1 and P27 | | Descriptor: | Cyclin-dependent kinase 4, Cyclin-dependent kinase inhibitor 1B, G1/S-specific cyclin-D1, ... | | Authors: | Guiley, K.Z, Stevenson, J.W, Lou, K, Barkovich, K.J, Bunch, K, Tripathi, S.M, Shokat, K.M, Rubin, S.M. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition.
Science, 366, 2019
|
|
5HZE
 
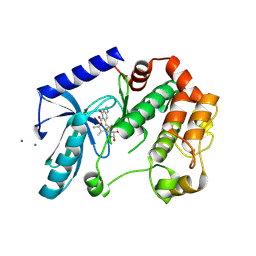 | | Mek1 adopts DFG-out conformation when bound to an analog of E6201. | | Descriptor: | (3S,4R,8S,9S,11E)-8,9,16-trihydroxy-3,4-dimethyl-14-(methylamino)-3,4,5,6,9,10-hexahydro-1H-2-benzoxacyclotetradecine-1,7(8H)-dione, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION | | Authors: | Larsen, N.A, Bloudoff, K. | | Deposit date: | 2016-02-02 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mek1 adopts DFG-out conformation when bound to an analog of E6201.
To Be Published
|
|
7KC5
 
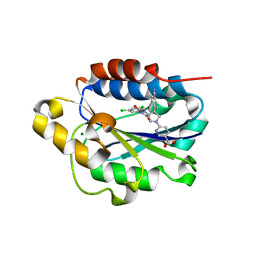 | | X-ray structure of Lfa-1 I domain in complex with BMS-68852 collected at 273 K | | Descriptor: | 6-[(5S,9R)-9-(4-cyanophenyl)-3-(3,5-dichlorophenyl)-1-methyl-2,4-dioxo-1,3,7-triazaspiro[4.4]non-7-yl]pyridine-3-carboxylic acid, Integrin alpha-L, MAGNESIUM ION | | Authors: | Woldeyes, R.A, Hallenbeck, K.K, Pfaff, S.J, Lee, G, Cortez, S.V, Kelly, M.J, Akassoglou, K, Arkin, M.R, Fraser, J.S. | | Deposit date: | 2020-10-05 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Divergent conformational dynamics controls allosteric ligand accessibility across evolutionarily related I-domain-containing integrins
To Be Published
|
|
7KC3
 
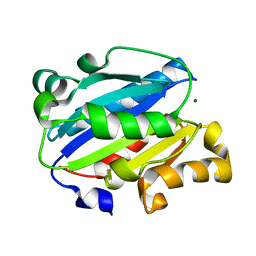 | | X-ray structure of Lfa-1 I domain collected at 273 K | | Descriptor: | Integrin alpha-L, MAGNESIUM ION | | Authors: | Woldeyes, R.A, Hallenbeck, K.K, Pfaff, S.J, Lee, G, Cortez, S.V, Kelly, M.J, Akassoglou, K, Arkin, M.R, Fraser, J.S. | | Deposit date: | 2020-10-05 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Divergent conformational dynamics controls allosteric ligand accessibility across evolutionarily related I-domain-containing integrins
To Be Published
|
|
7KC6
 
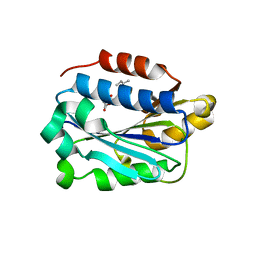 | | X-ray structure of Lfa-1 I domain in complex with Lovastatin collected at 273 K | | Descriptor: | Integrin alpha-L, LOVASTATIN, MAGNESIUM ION | | Authors: | Woldeyes, R.A, Hallenbeck, K.K, Pfaff, S.J, Lee, G, Cortez, S.V, Kelly, M.J, Akassoglou, K, Arkin, M.R, Fraser, J.S. | | Deposit date: | 2020-10-05 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Divergent conformational dynamics controls allosteric ligand accessibility across evolutionarily related I-domain-containing integrins
To Be Published
|
|
3FF3
 
 | |
2H5L
 
 | | S-Adenosylhomocysteine hydrolase containing NAD and 3-deaza-D-eritadenine | | Descriptor: | (2R,3R)-4-(4-AMINO-1H-IMIDAZO[4,5-C]PYRIDIN-1-YL)-2,3-DIHYDROXYBUTANOIC ACID, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, T, Komoto, J, Takusagawa, F. | | Deposit date: | 2006-05-26 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and function of eritadenine and its 3-deaza analogues: Potent inhibitors of S-adenosylhomocysteine hydrolase and hypocholesterolemic agents.
Biochem.Pharm., 73, 2007
|
|
2PVV
 
 | |
1CJP
 
 | | CONCANAVALIN A COMPLEX WITH 4'-METHYLUMBELLIFERYL-ALPHA-D-GLUCOPYRANOSIDE | | Descriptor: | 4-METHYLUMBELLIFERYL-ALPHA-D-GLUCOSE, CALCIUM ION, CONCANAVALIN A, ... | | Authors: | Hamodrakas, S.J, Kanellopoulos, P.N, Tucker, P.A. | | Deposit date: | 1996-10-03 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | The crystal structure of the complex of concanavalin A with 4'-methylumbelliferyl-alpha-D-glucopyranoside.
J.Struct.Biol., 118, 1997
|
|
4YJN
 
 | |
6N8G
 
 | | IRAK4 bound to benzoxazole compound | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-[2-(morpholin-4-yl)-6-(piperidin-1-yl)-1,3-benzoxazol-5-yl]-6-(1H-pyrrolo[2,3-b]pyridin-5-yl)pyridine-2-carboxamide | | Authors: | Larsen, N.A, Bloudoff, K, Subramanian, V, Dobrzanska, M, Gluza, K. | | Deposit date: | 2018-11-29 | | Release date: | 2018-12-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | To be published
To Be Published
|
|
4YSJ
 
 | |
8PI9
 
 | | DNA binding domain of HNF-1A bound to P2-HNF4A promoter DNA variant (P2 -181G>A) | | Descriptor: | Chains: E, Chains: F, Hepatocyte nuclear factor 1-alpha | | Authors: | Kind, L, Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular mechanism of HNF-1A-mediated HNF4A gene regulation and promoter-driven HNF4A-MODY diabetes.
JCI Insight, 9, 2024
|
|
8PI7
 
 | | DNA binding domain of HNF-1A bound to P2-HNF4A promoter DNA variant (P2 -169C>T) | | Descriptor: | CHLORIDE ION, Chains: E, Chains: F, ... | | Authors: | Kind, L, Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular mechanism of HNF-1A-mediated HNF4A gene regulation and promoter-driven HNF4A-MODY diabetes.
JCI Insight, 9, 2024
|
|
