7ZLY
 
 | | Crystal structure of human GPCR Niacin receptor (HCA2) expressed from Spodoptera frugiperda | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Hydroxycarboxylic acid receptor 2,Soluble cytochrome b562, OLEIC ACID | | Authors: | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, FiBerto, J.F, Wu, L.J, Tong, J.H, Han, G.W, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | Deposit date: | 2022-04-17 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
7ZL9
 
 | | Crystal structure of human GPCR Niacin receptor (HCA2) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DI(HYDROXYETHYL)ETHER, Hydroxycarboxylic acid receptor 2,Soluble cytochrome b562, ... | | Authors: | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, Han, G.W, FiBerto, J.F, Wu, L.J, Tong, J.H, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | Deposit date: | 2022-04-14 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
1SL8
 
 | | Calcium-loaded apo-aequorin from Aequorea victoria | | Descriptor: | Aequorin 1, CALCIUM ION | | Authors: | Deng, L, Markova, S.V, Vysotski, E.S, Liu, Z.J, Lee, J, Rose, J, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-05 | | Release date: | 2004-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | All three Ca2+-binding loops of photoproteins bind calcium ions: The crystal structures of calcium-loaded apo-aequorin and apo-obelin.
Protein Sci., 14, 2005
|
|
1SL7
 
 | | Crystal structure of calcium-loaded apo-obelin from Obelia longissima | | Descriptor: | CALCIUM ION, Obelin | | Authors: | Deng, L, Markova, S.V, Vysotski, E.S, Liu, Z.J, Lee, J, Rose, J, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-05 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | All three Ca2+-binding loops of photoproteins bind calcium ions: The crystal structures of calcium-loaded apo-aequorin and apo-obelin.
Protein Sci., 14, 2005
|
|
5TGZ
 
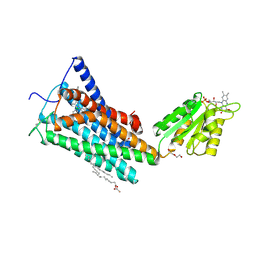 | | Crystal Structure of the Human Cannabinoid Receptor CB1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-[4-[2-(2,4-dichlorophenyl)-4-methyl-5-(piperidin-1-ylcarbamoyl)pyrazol-3-yl]phenyl]but-3-ynyl nitrate, Cannabinoid receptor 1,Flavodoxin,Cannabinoid receptor 1, ... | | Authors: | Hua, T, Vemuri, K, Pu, M, Qu, L, Han, G.W, Wu, Y, Zhao, S, Shui, W, Li, S, Korde, A, Laprairie, R.B, Stahl, E.L, Ho, J.H, Zvonok, N, Zhou, H, Kufareva, I, Wu, B, Zhao, Q, Hanson, M.A, Bohn, L.M, Makriyannis, A, Stevens, R.C, Liu, Z.J. | | Deposit date: | 2016-09-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Human Cannabinoid Receptor CB1.
Cell, 167, 2016
|
|
2O4X
 
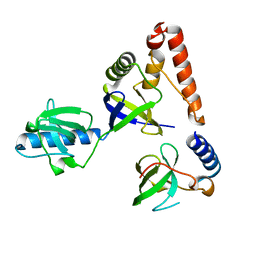 | | Crystal structure of human P100 tudor domain | | Descriptor: | Staphylococcal nuclease domain-containing protein 1 | | Authors: | Shaw, N, Zhao, M, Cheng, C, Xu, H, Yang, J, Silvennoinen, O, Rao, Z, Wang, B.C, Liu, Z.J. | | Deposit date: | 2006-12-05 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human P100 tudor domain
To be Published
|
|
2PW6
 
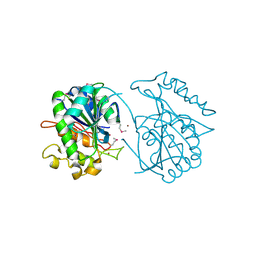 | | Crystal structure of uncharacterized protein JW3007 from Escherichia coli K12 | | Descriptor: | Uncharacterized protein ygiD, ZINC ION | | Authors: | Newton, M.G, Takagi, Y, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokayama, S, Li, Y, Chen, L, Zhu, J, Ruble, J, Liu, Z.J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-10 | | Release date: | 2007-06-12 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of uncharacterized protein JW3007 from Escherichia coli K12.
To be Published
|
|
2QVO
 
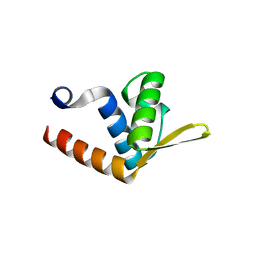 | | Crystal structure of AF1382 from Archaeoglobus fulgidus | | Descriptor: | Uncharacterized protein AF_1382 | | Authors: | Zhu, J, Zhao, M, Fu, Z.-Q, Yang, H, Chang, J, Hao, X, Chen, L, Liu, Z.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2007-08-08 | | Release date: | 2007-09-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of AF1382 from Archaeoglobus fulgidus.
To be Published
|
|
3LU1
 
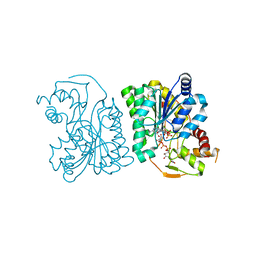 | | Crystal Structure Analysis of WbgU: a UDP-GalNAc 4-epimerase | | Descriptor: | GLYCINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Bhatt, V.S, Guo, C.Y, Zhao, G, Yi, W, Liu, Z.J, Wang, P.G. | | Deposit date: | 2010-02-16 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Altered architecture of substrate binding region defines the unique specificity of UDP-GalNAc 4-epimerases.
Protein Sci., 20, 2011
|
|
3CQ2
 
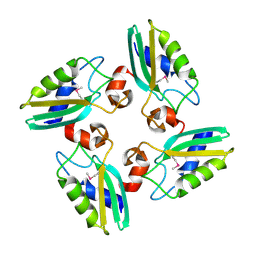 | | Structure of the DTDP-4-Keto-L-Rhamnose Reductase related protein (other form) from Thermus Thermophilus HB8 | | Descriptor: | Putative uncharacterized protein TTHB138 | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Satoh, S, Kitamura, Y, Ebihara, A, Chen, L, Liu, Z.J, Wang, B.C, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-02 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the DTDP-4-Keto-L-Rhamnose Reductase related protein from Thermus Thermophilus HB8
To be Published
|
|
3ECR
 
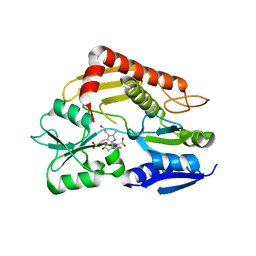 | | Structure of human porphobilinogen deaminase | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | Authors: | Song, G, Li, Y, Cheng, C, Zhao, Y, Gao, A, Zhang, R, Joachimiak, A, Shaw, N, Liu, Z.J. | | Deposit date: | 2008-09-01 | | Release date: | 2008-09-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Structural insight into acute intermittent porphyria.
Faseb J., 23, 2009
|
|
3FCX
 
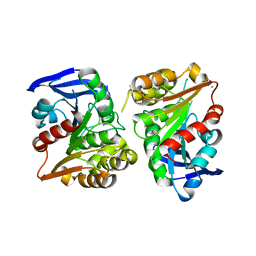 | | Crystal structure of human esterase D | | Descriptor: | CALCIUM ION, MAGNESIUM ION, S-formylglutathione hydrolase | | Authors: | Wu, D, Li, Y, Song, G, Zhang, D, Shaw, N, Liu, Z.J. | | Deposit date: | 2008-11-24 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human esterase D: a potential genetic marker of retinoblastoma
Faseb J., 23, 2009
|
|
3FHA
 
 | | Structure of endo-beta-N-acetylglucosaminidase A | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase, GLYCEROL, ... | | Authors: | Yin, J, Li, L, Shaw, N, Li, Y, Song, J.K, Zhang, W, Xia, C, Zhang, R, Joachimiak, A, Zhang, H.C, Wang, L.X, Wang, P, Liu, Z.J. | | Deposit date: | 2008-12-09 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis and catalytic mechanism for the dual functional endo-beta-N-acetylglucosaminidase A.
Plos One, 4, 2009
|
|
3GED
 
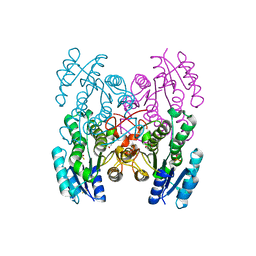 | | Fingerprint and Structural Analysis of a Apo SCOR enzyme from Clostridium thermocellum | | Descriptor: | GLYCEROL, SODIUM ION, Short-chain dehydrogenase/reductase SDR, ... | | Authors: | Huether, R, Liu, Z.J, Xu, H, Wang, B.C, Pletnev, V, Mao, Q, Umland, T, Duax, W. | | Deposit date: | 2009-02-25 | | Release date: | 2009-03-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Sequence fingerprint and structural analysis of the SCOR enzyme A3DFK9 from Clostridium thermocellum.
Proteins, 78, 2010
|
|
3GEG
 
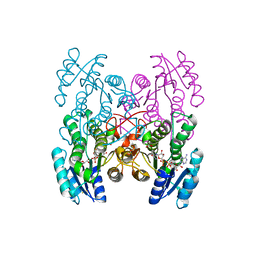 | | Fingerprint and Structural Analysis of a SCOR enzyme with its bound cofactor from Clostridium thermocellum | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Huether, R, Liu, Z.J, Xu, H, Wang, B.C, Pletnev, V, Mao, Q, Umland, T, Duax, W. | | Deposit date: | 2009-02-25 | | Release date: | 2009-03-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Sequence fingerprint and structural analysis of the SCOR enzyme A3DFK9 from Clostridium thermocellum.
Proteins, 78, 2010
|
|
1LKM
 
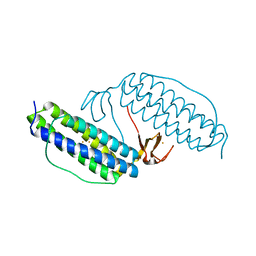 | | Crystal structure of Desulfovibrio vulgaris rubrerythrin all-iron(III) form | | Descriptor: | FE (III) ION, Rubrerythrin all-iron(III) form | | Authors: | Jin, S, Kurtz Jr, D.M, Liu, Z.J, Rose, J, Wang, B.C. | | Deposit date: | 2002-04-25 | | Release date: | 2002-09-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | X-ray Crystal Structures of Reduced Rubrerythrin and its Azide Adduct: A Structure-Based Mechanism for a Non-Heme DiIron Peroxidase
J.Am.Chem.Soc., 124, 2002
|
|
5H1Y
 
 | |
1LKO
 
 | | Crystal structure of Desulfovibrio vulgaris rubrerythrin all-iron(II) form | | Descriptor: | FE (II) ION, Rubrerythrin all-iron(II) form | | Authors: | Jin, S, Kurtz Jr, D.M, Liu, Z.J, Rose, J, Wang, B.C. | | Deposit date: | 2002-04-25 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray Crystal Structures of Reduced Rubrerythrin and its Azide Adduct: A Structure-Based Mechanism for a Non-Heme DiIron Peroxidase
J.Am.Chem.Soc., 124, 2002
|
|
1LKP
 
 | | Crystal structure of Desulfovibrio vulgaris rubrerythrin all-iron(II) form, azide adduct | | Descriptor: | AZIDE ION, FE (II) ION, Rubrerythrin | | Authors: | Jin, S, Kurtz Jr, D.M, Liu, Z.J, Rose, J, Wang, B.C. | | Deposit date: | 2002-04-25 | | Release date: | 2002-09-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | X-ray Crystal Structures of Reduced Rubrerythrin and its Azide Adduct: A Structure-Based Mechanism for a Non-Heme Diiron Peroxidase
J.Am.Chem.Soc., 124, 2002
|
|
3OVR
 
 | | Crystal Structure of hRPE and D-Xylulose 5-Phosphate Complex | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, 5-O-phosphono-D-xylulose, FE (II) ION, ... | | Authors: | Liang, W.G, Ouyang, S.Y, Shaw, N, Joachimiak, A, Zhang, R.G, Liu, Z.J. | | Deposit date: | 2010-09-17 | | Release date: | 2011-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Conversion of D-ribulose 5-phosphate to D-xylulose 5-phosphate: new insights from structural and biochemical studies on human RPE
Faseb J., 25, 2011
|
|
3OVP
 
 | | Crystal Structure of hRPE | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, FE (II) ION, Ribulose-phosphate 3-epimerase | | Authors: | Liang, W.G, Ouyang, S.Y, Shaw, N, Joachimiak, A, Zhang, R.G, Liu, Z.J. | | Deposit date: | 2010-09-16 | | Release date: | 2011-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Conversion of D-ribulose 5-phosphate to D-xylulose 5-phosphate: new insights from structural and biochemical studies on human RPE
Faseb J., 25, 2011
|
|
3OVQ
 
 | | Crystal Structure of hRPE and D-Ribulose-5-Phospate Complex | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, FE (II) ION, RIBULOSE-5-PHOSPHATE, ... | | Authors: | Liang, W.G, Ouyang, S.Y, Shaw, N, Joachimiak, A, Zhang, R.G, Liu, Z.J. | | Deposit date: | 2010-09-17 | | Release date: | 2011-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Conversion of D-ribulose 5-phosphate to D-xylulose 5-phosphate: new insights from structural and biochemical studies on human RPE
Faseb J., 25, 2011
|
|
1VK1
 
 | | Conserved hypothetical protein from Pyrococcus furiosus Pfu-392566-001 | | Descriptor: | Conserved hypothetical protein, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Shah, A, Liu, Z.J, Tempel, W, Chen, L, Lee, D, Yang, H, Chang, J, Zhao, M, Ng, J, Rose, J, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-04-13 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | (NZ)CH...O contacts assist crystallization of a ParB-like nuclease.
Bmc Struct.Biol., 7, 2007
|
|
8WRZ
 
 | | Cry-EM structure of cannabinoid receptor-arrestin 2 complex | | Descriptor: | (6~{a}~{R},9~{R},10~{a}~{R})-9-(hydroxymethyl)-3-(8-isothiocyanato-2-methyl-octan-2-yl)-6,6-dimethyl-6~{a},7,8,9,10,10~{a}-hexahydrobenzo[c]chromen-1-ol, Beta-arrestin-1, Soluble cytochrome b562,Cannabinoid receptor 1, ... | | Authors: | Wang, Y.X, Wang, T, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2023-10-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of cannabinoid receptor CB1-beta-arrestin complex.
Protein Cell, 15, 2024
|
|
1QYB
 
 | | X-ray crystal structure of Desulfovibrio vulgaris rubrerythrin with zinc substituted into the [Fe(SCys)4] site and alternative diiron site structures | | Descriptor: | FE (III) ION, Rubrerythrin, SULFATE ION, ... | | Authors: | Jin, S, Kurtz, D.M, Liu, Z.J, Rose, J, Wang, B.C. | | Deposit date: | 2003-09-10 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | X-ray Crystal Structure of Desulfovibrio vulgaris Rubrerythrin with Zinc Substituted into the [Fe(SCys)(4)] Site and Alternative Diiron Site Structures.
Biochemistry, 43, 2004
|
|
