2MAI
 
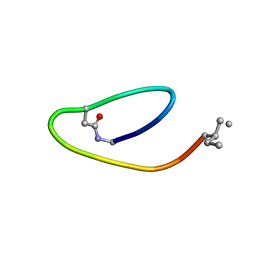 | | NMR structure of lassomycin | | Descriptor: | Lassomycin | | Authors: | Gavrish, E, Sit, C.S, Kandror, O, Spoering, A, Peoples, A, Ling, L, Fetterman, A, Hughes, D, Cao, S, Bissell, A, Torrey, H, Akopian, T, Mueller, A, Epstein, S, Goldberg, A, Clardy, J, Lewis, K. | | Deposit date: | 2013-07-09 | | Release date: | 2014-05-07 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Lassomycin, a Ribosomally Synthesized Cyclic Peptide, Kills Mycobacterium tuberculosis by Targeting the ATP-Dependent Protease ClpC1P1P2.
Chem.Biol., 21, 2014
|
|
7L0R
 
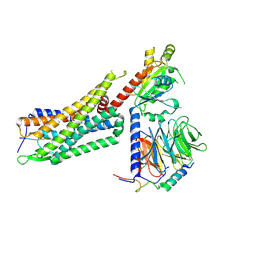 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, noncanonical state, without AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7L0P
 
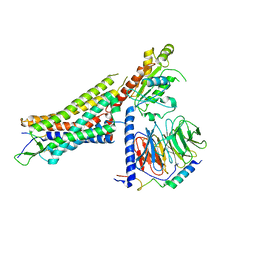 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, canonical state, without AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7L0S
 
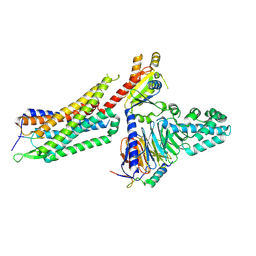 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, noncanonical state, with AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7L0Q
 
 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, canonical state, with AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6Z66
 
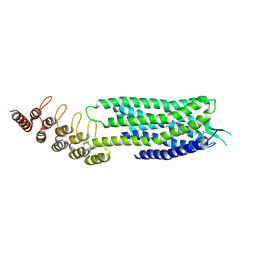 | | Crystal structure of apo-state neurotensin receptor 1 | | Descriptor: | Neurotensin receptor type 1,Neurotensin receptor type 1,DARPin | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-05-27 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.192 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6ZA8
 
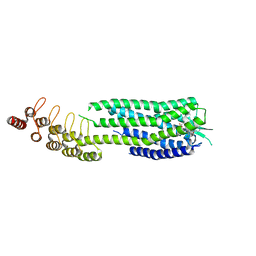 | | Crystal structure of the neurotensin receptor 1 in complex with the small-molecule partial agonist RTI-3a | | Descriptor: | (2~{S})-2-[[1-(7-chloranylquinolin-4-yl)-5-(2,6-dimethoxyphenyl)pyrazol-3-yl]carbonylamino]-4-methyl-pentanoic acid, Neurotensin receptor type 1,Neurotensin receptor type 1,Neurotensin receptor 1 (NTSR1),Neurotensin receptor 1 (NTSR1),Neurotensin receptor type 1,Neurotensin receptor 1 (NTSR1),Neurotensin receptor 1 (NTSR1) | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-06-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6Z4V
 
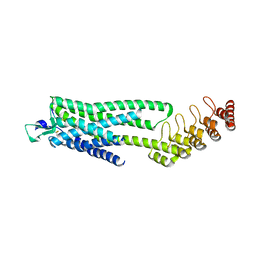 | | Crystal structure of the neurotensin receptor 1 (NTSR1-H4bmx) in complex with NTS8-13 | | Descriptor: | ARG-PRO-TYR-ILE-LEU, Neurotensin receptor type 1,Neurotensin receptor type 1,DARPin | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-05-25 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6Z4Q
 
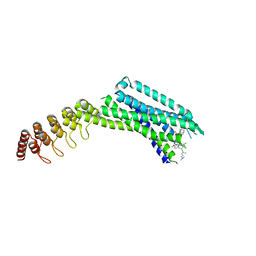 | | Crystal structure of the neurotensin receptor 1 in complex with the small-molecule inverse agonist SR142948A | | Descriptor: | 2-[[5-(2,6-dimethoxyphenyl)-1-[4-[3-(dimethylamino)propyl-methyl-carbamoyl]-2-propan-2-yl-phenyl]pyrazol-3-yl]carbonylamino]adamantane-2-carboxylic acid, Neurotensin receptor type 1,Neurotensin receptor type 1,DARPin | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-05-25 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.923 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6ZIN
 
 | | Crystal structure of the neurotensin receptor 1 in complex with the small molecule inverse agonist SR48692 | | Descriptor: | 2-[[1-(7-chloranylquinolin-4-yl)-5-(2,6-dimethoxyphenyl)pyrazol-3-yl]carbonylamino]adamantane-2-carboxylic acid, Neurotensin receptor type 1,DARPin,HRV 3C protease recognition sequence | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-06-26 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.639 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6Z8N
 
 | | Crystal structure of the neurotensin receptor 1 in complex with the small-molecule full agonist SRI-9829 | | Descriptor: | (2~{S})-4-methyl-2-[(1-quinolin-8-ylsulfonylindol-3-yl)carbonylamino]pentanoic acid, Neurotensin receptor type 1,Neurotensin receptor type 1,Neurotensin receptor type 1,Neurotensin receptor type 1,Neurotensin receptor 1 (NTSR1),Neurotensin receptor 1 (NTSR1) | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-06-02 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6YVR
 
 | | Crystal structure of the neurotensin receptor 1 in complex with the peptide full agonist NTS8-13 | | Descriptor: | Neurotensin receptor type 1,Neurotensin receptor type 1,DARPin crystallisation chaperone, neurotensin NTS8-13 (full agonist), nonyl beta-D-glucopyranoside | | Authors: | Deluigi, M, Merklinger, L, Hilge, M, Ernst, P, Klipp, A, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-04-28 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.458 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6Z4S
 
 | | Crystal structure of the neurotensin receptor 1 (NTSR1-H4bmx) in complex with the small molecule inverse agonist SR48692 | | Descriptor: | 2-[[1-(7-chloranylquinolin-4-yl)-5-(2,6-dimethoxyphenyl)pyrazol-3-yl]carbonylamino]adamantane-2-carboxylic acid, Neurotensin receptor type 1,DARPin,HRV 3C protease recognition sequence | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-05-25 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.707 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
7B6W
 
 | | Crystal structure of the human alpha1B adrenergic receptor in complex with inverse agonist (+)-cyclazosin | | Descriptor: | Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor, [(4~{a}~{R},8~{a}~{S})-4-(4-azanyl-6,7-dimethoxy-quinazolin-2-yl)-2,3,4~{a},5,6,7,8,8~{a}-octahydroquinoxalin-1-yl]-(furan-2-yl)methanone | | Authors: | Deluigi, M, Morstein, L, Hilge, M, Schuster, M, Merklinger, L, Klipp, A, Scott, D.J, Plueckthun, A. | | Deposit date: | 2020-12-08 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.873 Å) | | Cite: | Crystal structure of the alpha 1B -adrenergic receptor reveals molecular determinants of selective ligand recognition.
Nat Commun, 13, 2022
|
|
5JLT
 
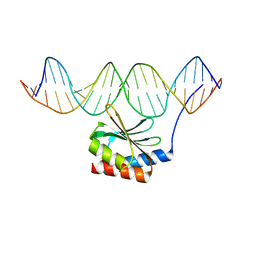 | | The crystal structure of the bacteriophage T4 MotA C-terminal domain in complex with dsDNA reveals a novel protein-DNA recognition motif | | Descriptor: | DNA (5'-D(*GP*AP*AP*GP*CP*TP*TP*TP*GP*CP*TP*TP*AP*AP*TP*AP*AP*TP*CP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*GP*AP*TP*TP*AP*TP*TP*AP*AP*GP*CP*AP*AP*AP*GP*CP*TP*TP*C)-3'), Middle transcription regulatory protein motA | | Authors: | Cuypers, M.G, Robertson, R.M, Knipling, L, Hinton, D.M, White, S.W. | | Deposit date: | 2016-04-27 | | Release date: | 2017-05-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | The phage T4 MotA transcription factor contains a novel DNA binding motif that specifically recognizes modified DNA.
Nucleic Acids Res., 46, 2018
|
|
4QCJ
 
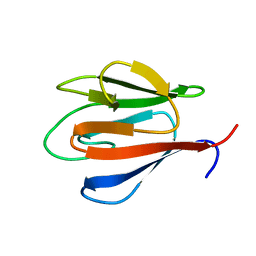 | | Crystal Structure of OdhI from Corynebacterium glutamicum | | Descriptor: | Oxoglutarate dehydrogenase inhibitor | | Authors: | Labahn, J, Raasch, K, Eggeling, L, Bocola, M, Leitner, A, Bott, M. | | Deposit date: | 2014-05-12 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interaction of 2-oxoglutarate dehydrogenase OdhA with its inhibitor OdhI in Corynebacterium glutamicum: Mutants and a model.
J.Biotechnol., 191, 2014
|
|
4MAD
 
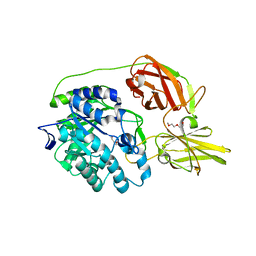 | | Crystal structure of beta-galactosidase C (BgaC) from Bacillus circulans ATCC 31382 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Beta-galactosidase | | Authors: | Kamerke, C, You, D.J, Kanaya, S, Elling, L. | | Deposit date: | 2013-08-16 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational design of a glycosynthase by the crystal structure of beta-galactosidase from Bacillus circulans (BgaC) and its use for the synthesis of N-acetyllactosamine type 1 glycan structures.
J.Biotechnol., 191, 2014
|
|
3R21
 
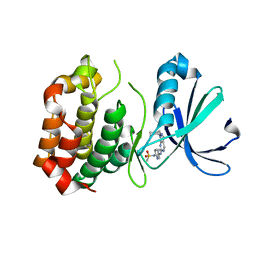 | | Design, synthesis, and biological evaluation of pyrazolopyridine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (Part I) | | Descriptor: | MAGNESIUM ION, N-(2-aminoethyl)-N-{5-[(1-cycloheptyl-1H-pyrazolo[3,4-d]pyrimidin-6-yl)amino]pyridin-2-yl}methanesulfonamide, Serine/threonine-protein kinase 6 | | Authors: | Zhang, L, Fan, J, Chong, J.-H, Cesena, A, Tam, B, Gilson, C, Boykin, C, Wang, D, Marcotte, D, Le Brazidec, J.-Y, Aivazian, D, Piao, J, Lundgren, K, Hong, K, Vu, K, Nguyen, K. | | Deposit date: | 2011-03-11 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, synthesis, and biological evaluation of pyrazolopyrimidine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (part I).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3KCF
 
 | |
4DHF
 
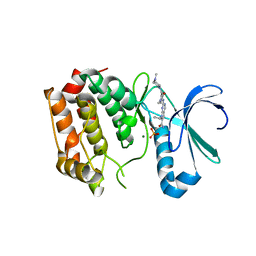 | | Structure of Aurora A mutant bound to Biogenidec cpd 15 | | Descriptor: | 7-cyclopentyl-2-({1-methyl-5-[(4-methylpiperazin-1-yl)carbonyl]-1H-pyrrol-3-yl}amino)-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Aurora kinase A, MAGNESIUM ION, ... | | Authors: | Silvian, L, Marcotte, D.J. | | Deposit date: | 2012-01-27 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of 2,6,7-trisubstituted-7H-pyrrolo[2,3-d]pyrimidines as Aurora kinases inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3R22
 
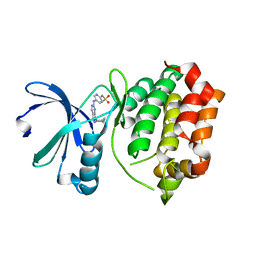 | | Design, synthesis, and biological evaluation of pyrazolopyridine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (Part I) | | Descriptor: | N-{5-[(1-cycloheptyl-1H-pyrazolo[3,4-d]pyrimidin-6-yl)amino]pyridin-2-yl}methanesulfonamide, Serine/threonine-protein kinase 6 | | Authors: | Zhang, L, Fan, J, Chong, J.-H, Cesana, A, Tam, B, Gilson, C, Boykin, C, Wang, D, Marcotte, D, Le Brazidec, J.-Y, Aivazian, D, Piao, J, Lundgren, K, Hong, K, Vu, K, Nguyen, K. | | Deposit date: | 2011-03-11 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, synthesis, and biological evaluation of pyrazolopyrimidine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (part I).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3FAA
 
 | |
8K8T
 
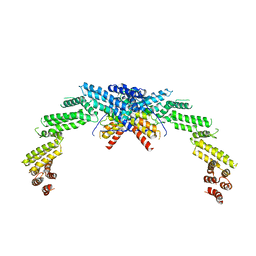 | | Structure of CUL3-RBX1-KLHL22 complex | | Descriptor: | Cullin-3, Kelch-like protein 22 | | Authors: | Wang, W, Ling, L, Dai, Z, Zuo, P, Yin, Y. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A conserved N-terminal motif of CUL3 contributes to assembly and E3 ligase activity of CRL3 KLHL22.
Nat Commun, 15, 2024
|
|
8K9I
 
 | | Structure of CUL3-RBX1-KLHL22 complex without CUL3 NA motif | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Wang, W, Ling, L, Dai, Z, Zuo, P, Yin, Y. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A conserved N-terminal motif of CUL3 contributes to assembly and E3 ligase activity of CRL3 KLHL22.
Nat Commun, 15, 2024
|
|
4CTB
 
 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with the inhibitor (5R)-8-amino-3-fluoro-5,19-dimethyl-20-oxo-5,18,19,20- tetrahydro-7,11-(azeno)pyrido(2',1':2,3)imidazo(4,5-h)(2,5,11) benzoxadiazacyclotetradecine-14-carbonitrile | | Descriptor: | (5R)-8-amino-3-fluoro-5,19-dimethyl-20-oxo-5,18,19,20-tetrahydro-11,7-(azeno)pyrido[2',1':2,3]imidazo[4,5-h][2,5,11]benzoxadiazacyclotetradecine-14-carbonitrile, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M.A, Deng, Y.L, Liu, W, Brooun, A, Stewart, A.E. | | Deposit date: | 2014-03-12 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of (10R)-7-Amino-12-Fluoro-2,10,16-Trimethyl-15-Oxo-10,15,16,17-Tetrahydro-2H-8,4-(Metheno)Pyrazolo[4,3-H][2,5,11]Benzoxadiazacyclotetradecine-3-Carbonitrile (Pf-06463922), a Macrocyclic Inhibitor of Alk/Ros1 with Pre-Clinical Brain Exposure and Broad Spectrum Potency Against Alk-Resistant Mutations.
J.Med.Chem., 57, 2014
|
|
